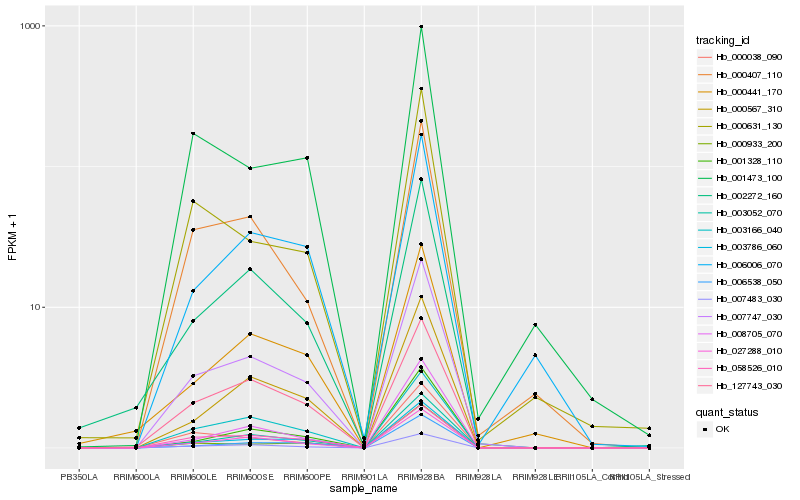
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007747_030 |
0.0 |
- |
- |
PREDICTED: WD repeat-containing protein LWD2-like [Jatropha curcas] |
| 2 |
Hb_007483_030 |
0.0594559566 |
- |
- |
- |
| 3 |
Hb_027288_010 |
0.0609466251 |
- |
- |
mutt domain protein, putative [Ricinus communis] |
| 4 |
Hb_003052_070 |
0.0632918143 |
- |
- |
PREDICTED: peroxidase 11 [Jatropha curcas] |
| 5 |
Hb_003786_060 |
0.0731649762 |
- |
- |
PREDICTED: transmembrane protein 220 [Jatropha curcas] |
| 6 |
Hb_003166_040 |
0.0853319157 |
- |
- |
PREDICTED: probable indole-3-acetic acid-amido synthetase GH3.1 [Jatropha curcas] |
| 7 |
Hb_058526_010 |
0.091184317 |
- |
- |
- |
| 8 |
Hb_000567_310 |
0.0923132808 |
- |
- |
Miraculin precursor, putative [Ricinus communis] |
| 9 |
Hb_002272_160 |
0.0957349437 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 10 |
Hb_006538_050 |
0.1001735947 |
- |
- |
hypothetical protein RCOM_1308310 [Ricinus communis] |
| 11 |
Hb_127743_030 |
0.1005042387 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 12 |
Hb_001328_110 |
0.1008509034 |
- |
- |
PREDICTED: LON peptidase N-terminal domain and RING finger protein 1-like [Jatropha curcas] |
| 13 |
Hb_000407_110 |
0.1022726716 |
- |
- |
ripening-induced ACC oxidase [Carica papaya] |
| 14 |
Hb_008705_070 |
0.1092417689 |
- |
- |
PREDICTED: uncharacterized protein LOC105638774 [Jatropha curcas] |
| 15 |
Hb_000038_090 |
0.112469419 |
transcription factor |
TF Family: NAC |
hypothetical protein POPTR_0002s05820g [Populus trichocarpa] |
| 16 |
Hb_001473_100 |
0.11375417 |
- |
- |
chalcone synthase 1 [Jatropha curcas] |
| 17 |
Hb_000933_200 |
0.1204466252 |
- |
- |
Cationic peroxidase 2 precursor, putative [Ricinus communis] |
| 18 |
Hb_000441_170 |
0.1214437307 |
- |
- |
PREDICTED: uncharacterized protein LOC105636851 [Jatropha curcas] |
| 19 |
Hb_000631_130 |
0.1220495708 |
- |
- |
PREDICTED: flavonoid 3'-monooxygenase [Jatropha curcas] |
| 20 |
Hb_006006_070 |
0.1228543741 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |