| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
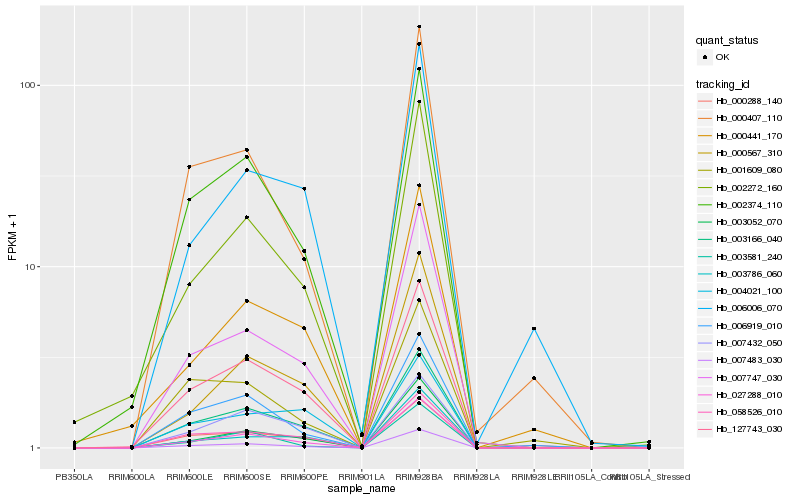
Hb_003166_040 |
0.0 |
- |
- |
PREDICTED: probable indole-3-acetic acid-amido synthetase GH3.1 [Jatropha curcas] |
| 2 |
Hb_127743_030 |
0.0296732016 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 3 |
Hb_058526_010 |
0.0471551186 |
- |
- |
- |
| 4 |
Hb_027288_010 |
0.0689059766 |
- |
- |
mutt domain protein, putative [Ricinus communis] |
| 5 |
Hb_007483_030 |
0.0744931221 |
- |
- |
- |
| 6 |
Hb_002374_110 |
0.0756796689 |
- |
- |
PREDICTED: uncharacterized protein LOC105638965 [Jatropha curcas] |
| 7 |
Hb_007432_050 |
0.0813156537 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB108 [Jatropha curcas] |
| 8 |
Hb_007747_030 |
0.0853319157 |
- |
- |
PREDICTED: WD repeat-containing protein LWD2-like [Jatropha curcas] |
| 9 |
Hb_003052_070 |
0.1048431142 |
- |
- |
PREDICTED: peroxidase 11 [Jatropha curcas] |
| 10 |
Hb_002272_160 |
0.1069250288 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 11 |
Hb_006919_010 |
0.107021809 |
- |
- |
PREDICTED: putative wall-associated receptor kinase-like 16 [Vitis vinifera] |
| 12 |
Hb_000567_310 |
0.1098109645 |
- |
- |
Miraculin precursor, putative [Ricinus communis] |
| 13 |
Hb_004021_100 |
0.1117839291 |
- |
- |
- |
| 14 |
Hb_000407_110 |
0.1121757381 |
- |
- |
ripening-induced ACC oxidase [Carica papaya] |
| 15 |
Hb_000288_140 |
0.1143237364 |
- |
- |
PREDICTED: putative cysteine-rich receptor-like protein kinase 35 [Jatropha curcas] |
| 16 |
Hb_003786_060 |
0.1146969776 |
- |
- |
PREDICTED: transmembrane protein 220 [Jatropha curcas] |
| 17 |
Hb_000441_170 |
0.1265657404 |
- |
- |
PREDICTED: uncharacterized protein LOC105636851 [Jatropha curcas] |
| 18 |
Hb_006006_070 |
0.1276682656 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 19 |
Hb_001609_080 |
0.1288392694 |
- |
- |
hypothetical protein POPTR_0017s05660g [Populus trichocarpa] |
| 20 |
Hb_003581_240 |
0.1305889576 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |