| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
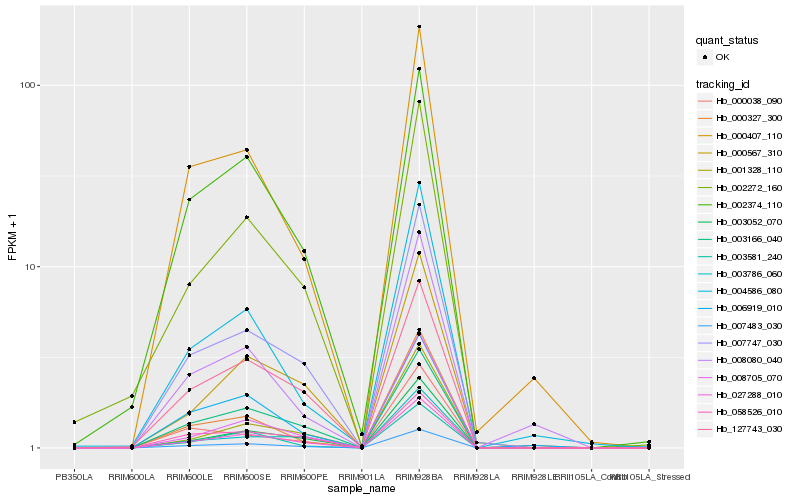
Hb_007483_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_027288_010 |
0.0318425814 |
- |
- |
mutt domain protein, putative [Ricinus communis] |
| 3 |
Hb_007747_030 |
0.0594559566 |
- |
- |
PREDICTED: WD repeat-containing protein LWD2-like [Jatropha curcas] |
| 4 |
Hb_003052_070 |
0.0719402312 |
- |
- |
PREDICTED: peroxidase 11 [Jatropha curcas] |
| 5 |
Hb_003166_040 |
0.0744931221 |
- |
- |
PREDICTED: probable indole-3-acetic acid-amido synthetase GH3.1 [Jatropha curcas] |
| 6 |
Hb_000407_110 |
0.0758735851 |
- |
- |
ripening-induced ACC oxidase [Carica papaya] |
| 7 |
Hb_002272_160 |
0.0903458458 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 8 |
Hb_127743_030 |
0.093002929 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 9 |
Hb_003581_240 |
0.093006713 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 10 |
Hb_058526_010 |
0.0951073061 |
- |
- |
- |
| 11 |
Hb_004586_080 |
0.0963979401 |
- |
- |
hypothetical protein POPTR_0002s10520g [Populus trichocarpa] |
| 12 |
Hb_002374_110 |
0.1005585406 |
- |
- |
PREDICTED: uncharacterized protein LOC105638965 [Jatropha curcas] |
| 13 |
Hb_006919_010 |
0.1014425255 |
- |
- |
PREDICTED: putative wall-associated receptor kinase-like 16 [Vitis vinifera] |
| 14 |
Hb_000567_310 |
0.101739817 |
- |
- |
Miraculin precursor, putative [Ricinus communis] |
| 15 |
Hb_000327_300 |
0.1079623924 |
- |
- |
PREDICTED: ABC transporter G family member 6-like [Jatropha curcas] |
| 16 |
Hb_008080_040 |
0.1085846207 |
- |
- |
hypothetical protein EUGRSUZ_D00834 [Eucalyptus grandis] |
| 17 |
Hb_008705_070 |
0.1090363056 |
- |
- |
PREDICTED: uncharacterized protein LOC105638774 [Jatropha curcas] |
| 18 |
Hb_003786_060 |
0.1095558911 |
- |
- |
PREDICTED: transmembrane protein 220 [Jatropha curcas] |
| 19 |
Hb_001328_110 |
0.1132148346 |
- |
- |
PREDICTED: LON peptidase N-terminal domain and RING finger protein 1-like [Jatropha curcas] |
| 20 |
Hb_000038_090 |
0.1137521483 |
transcription factor |
TF Family: NAC |
hypothetical protein POPTR_0002s05820g [Populus trichocarpa] |