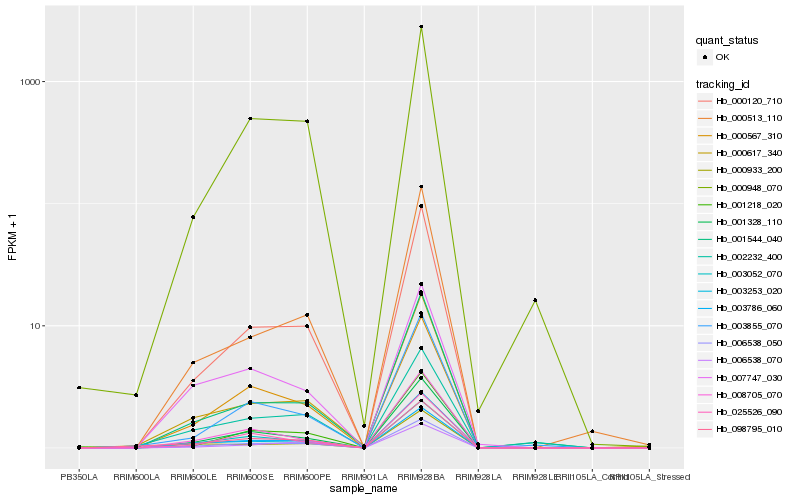
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006538_050 |
0.0 |
- |
- |
hypothetical protein RCOM_1308310 [Ricinus communis] |
| 2 |
Hb_003786_060 |
0.0503514943 |
- |
- |
PREDICTED: transmembrane protein 220 [Jatropha curcas] |
| 3 |
Hb_000120_710 |
0.054698477 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 4 |
Hb_001328_110 |
0.0569230022 |
- |
- |
PREDICTED: LON peptidase N-terminal domain and RING finger protein 1-like [Jatropha curcas] |
| 5 |
Hb_003052_070 |
0.0637103504 |
- |
- |
PREDICTED: peroxidase 11 [Jatropha curcas] |
| 6 |
Hb_000567_310 |
0.0676806272 |
- |
- |
Miraculin precursor, putative [Ricinus communis] |
| 7 |
Hb_000617_340 |
0.0841330772 |
- |
- |
respiratory burst oxidase B [Manihot esculenta] |
| 8 |
Hb_000948_070 |
0.0894543946 |
- |
- |
hypothetical protein POPTR_0016s01720g [Populus trichocarpa] |
| 9 |
Hb_003253_020 |
0.0930646185 |
transcription factor |
TF Family: B3 |
transcription factor, putative [Ricinus communis] |
| 10 |
Hb_008705_070 |
0.0931632159 |
- |
- |
PREDICTED: uncharacterized protein LOC105638774 [Jatropha curcas] |
| 11 |
Hb_001544_040 |
0.0938833607 |
- |
- |
PREDICTED: uncharacterized protein At1g66480-like [Populus euphratica] |
| 12 |
Hb_001218_020 |
0.0977858393 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0005s17080g [Populus trichocarpa] |
| 13 |
Hb_003855_070 |
0.0981115518 |
- |
- |
hypothetical protein POPTR_0001s30980g [Populus trichocarpa] |
| 14 |
Hb_000933_200 |
0.0995062476 |
- |
- |
Cationic peroxidase 2 precursor, putative [Ricinus communis] |
| 15 |
Hb_006538_070 |
0.0995315147 |
- |
- |
hypothetical protein JCGZ_18339 [Jatropha curcas] |
| 16 |
Hb_007747_030 |
0.1001735947 |
- |
- |
PREDICTED: WD repeat-containing protein LWD2-like [Jatropha curcas] |
| 17 |
Hb_098795_010 |
0.1013611808 |
- |
- |
PREDICTED: naringenin,2-oxoglutarate 3-dioxygenase-like [Jatropha curcas] |
| 18 |
Hb_025526_090 |
0.1055376754 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g27290 [Jatropha curcas] |
| 19 |
Hb_002232_400 |
0.1081625568 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH25-like [Jatropha curcas] |
| 20 |
Hb_000513_110 |
0.1083401453 |
- |
- |
PREDICTED: germin-like protein subfamily 1 member 13 [Populus euphratica] |