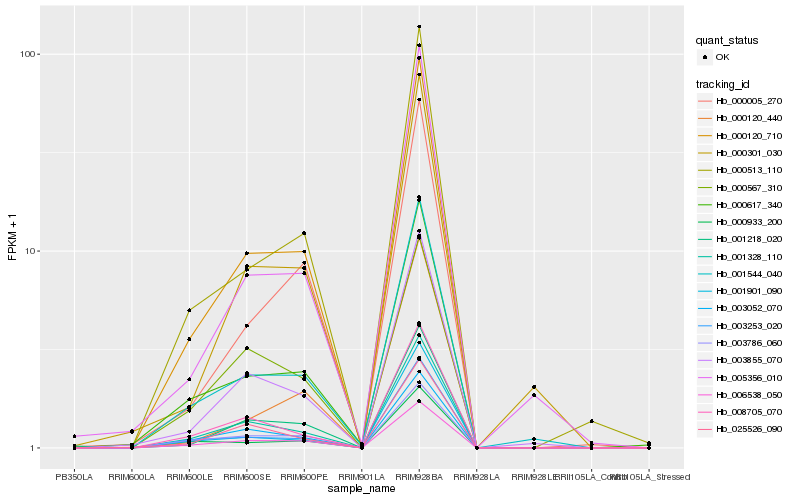
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000120_710 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_006538_050 |
0.054698477 |
- |
- |
hypothetical protein RCOM_1308310 [Ricinus communis] |
| 3 |
Hb_001328_110 |
0.0601811029 |
- |
- |
PREDICTED: LON peptidase N-terminal domain and RING finger protein 1-like [Jatropha curcas] |
| 4 |
Hb_001544_040 |
0.067670411 |
- |
- |
PREDICTED: uncharacterized protein At1g66480-like [Populus euphratica] |
| 5 |
Hb_000617_340 |
0.0684271611 |
- |
- |
respiratory burst oxidase B [Manihot esculenta] |
| 6 |
Hb_000513_110 |
0.0695475593 |
- |
- |
PREDICTED: germin-like protein subfamily 1 member 13 [Populus euphratica] |
| 7 |
Hb_003253_020 |
0.0737371839 |
transcription factor |
TF Family: B3 |
transcription factor, putative [Ricinus communis] |
| 8 |
Hb_003855_070 |
0.0756574019 |
- |
- |
hypothetical protein POPTR_0001s30980g [Populus trichocarpa] |
| 9 |
Hb_001218_020 |
0.0784221477 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0005s17080g [Populus trichocarpa] |
| 10 |
Hb_008705_070 |
0.0883802138 |
- |
- |
PREDICTED: uncharacterized protein LOC105638774 [Jatropha curcas] |
| 11 |
Hb_000005_270 |
0.0897743074 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase protein 2 precursor, putative [Ricinus communis] |
| 12 |
Hb_000933_200 |
0.0973686613 |
- |
- |
Cationic peroxidase 2 precursor, putative [Ricinus communis] |
| 13 |
Hb_001901_090 |
0.0986115725 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 14 |
Hb_003786_060 |
0.099287248 |
- |
- |
PREDICTED: transmembrane protein 220 [Jatropha curcas] |
| 15 |
Hb_003052_070 |
0.10027177 |
- |
- |
PREDICTED: peroxidase 11 [Jatropha curcas] |
| 16 |
Hb_000301_030 |
0.1012907686 |
- |
- |
PREDICTED: pectate lyase-like [Jatropha curcas] |
| 17 |
Hb_005356_010 |
0.1043839628 |
- |
- |
hypothetical protein POPTR_0003s16550g [Populus trichocarpa] |
| 18 |
Hb_025526_090 |
0.1067974726 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g27290 [Jatropha curcas] |
| 19 |
Hb_000120_440 |
0.10745281 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000567_310 |
0.1084336099 |
- |
- |
Miraculin precursor, putative [Ricinus communis] |