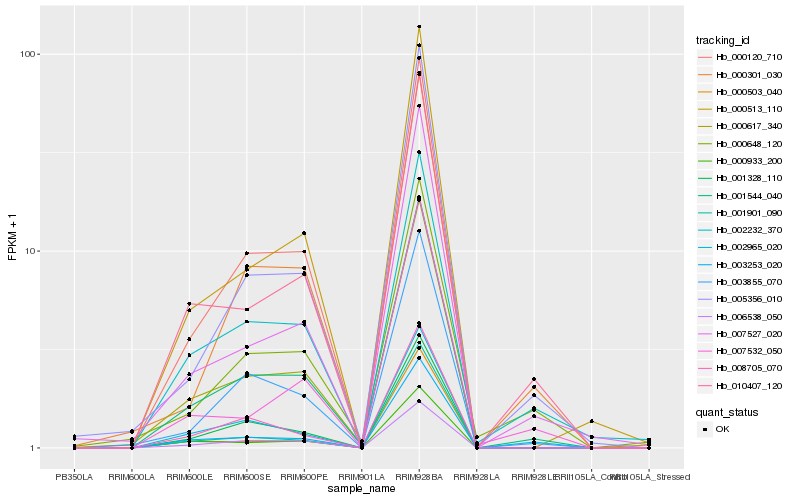
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001544_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At1g66480-like [Populus euphratica] |
| 2 |
Hb_000120_710 |
0.067670411 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 3 |
Hb_010407_120 |
0.0696849848 |
- |
- |
PREDICTED: glutathione transferase GST 23-like [Jatropha curcas] |
| 4 |
Hb_003253_020 |
0.0696860321 |
transcription factor |
TF Family: B3 |
transcription factor, putative [Ricinus communis] |
| 5 |
Hb_000617_340 |
0.0708249756 |
- |
- |
respiratory burst oxidase B [Manihot esculenta] |
| 6 |
Hb_000513_110 |
0.0786543907 |
- |
- |
PREDICTED: germin-like protein subfamily 1 member 13 [Populus euphratica] |
| 7 |
Hb_005356_010 |
0.0808766285 |
- |
- |
hypothetical protein POPTR_0003s16550g [Populus trichocarpa] |
| 8 |
Hb_003855_070 |
0.0812692771 |
- |
- |
hypothetical protein POPTR_0001s30980g [Populus trichocarpa] |
| 9 |
Hb_007527_020 |
0.084874529 |
- |
- |
PREDICTED: feruloyl CoA ortho-hydroxylase 1 [Jatropha curcas] |
| 10 |
Hb_001328_110 |
0.0871803948 |
- |
- |
PREDICTED: LON peptidase N-terminal domain and RING finger protein 1-like [Jatropha curcas] |
| 11 |
Hb_001901_090 |
0.0872488742 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 12 |
Hb_000933_200 |
0.0921761274 |
- |
- |
Cationic peroxidase 2 precursor, putative [Ricinus communis] |
| 13 |
Hb_006538_050 |
0.0938833607 |
- |
- |
hypothetical protein RCOM_1308310 [Ricinus communis] |
| 14 |
Hb_002965_020 |
0.0940079491 |
- |
- |
hypothetical protein JCGZ_18947 [Jatropha curcas] |
| 15 |
Hb_000301_030 |
0.0948682365 |
- |
- |
PREDICTED: pectate lyase-like [Jatropha curcas] |
| 16 |
Hb_008705_070 |
0.0986902885 |
- |
- |
PREDICTED: uncharacterized protein LOC105638774 [Jatropha curcas] |
| 17 |
Hb_002232_370 |
0.103716532 |
- |
- |
PREDICTED: receptor-like protein kinase HAIKU2 [Jatropha curcas] |
| 18 |
Hb_000503_040 |
0.106716167 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase-like [Jatropha curcas] |
| 19 |
Hb_000648_120 |
0.1070481422 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase IV.1 [Vitis vinifera] |
| 20 |
Hb_007532_050 |
0.1073653102 |
- |
- |
9-cis-epoxycarotenoid dioxygenase, putative [Ricinus communis] |