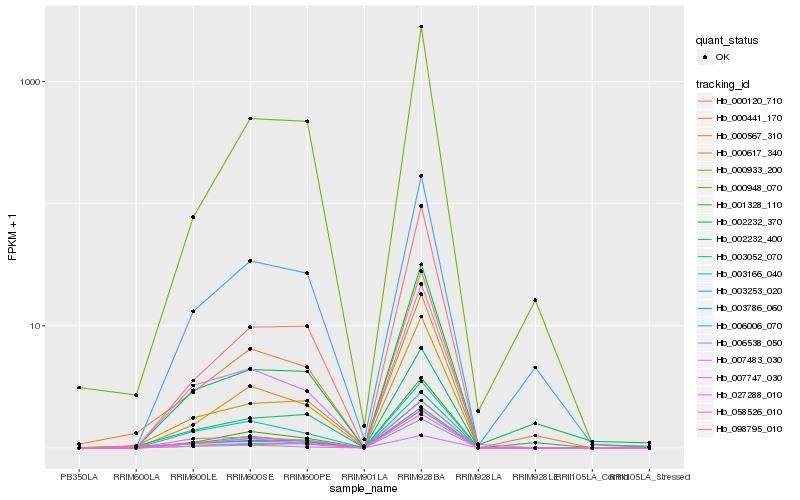
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003786_060 |
0.0 |
- |
- |
PREDICTED: transmembrane protein 220 [Jatropha curcas] |
| 2 |
Hb_006538_050 |
0.0503514943 |
- |
- |
hypothetical protein RCOM_1308310 [Ricinus communis] |
| 3 |
Hb_003052_070 |
0.0660679902 |
- |
- |
PREDICTED: peroxidase 11 [Jatropha curcas] |
| 4 |
Hb_007747_030 |
0.0731649762 |
- |
- |
PREDICTED: WD repeat-containing protein LWD2-like [Jatropha curcas] |
| 5 |
Hb_000567_310 |
0.0793259523 |
- |
- |
Miraculin precursor, putative [Ricinus communis] |
| 6 |
Hb_001328_110 |
0.0890333247 |
- |
- |
PREDICTED: LON peptidase N-terminal domain and RING finger protein 1-like [Jatropha curcas] |
| 7 |
Hb_000933_200 |
0.0960674364 |
- |
- |
Cationic peroxidase 2 precursor, putative [Ricinus communis] |
| 8 |
Hb_027288_010 |
0.0987532025 |
- |
- |
mutt domain protein, putative [Ricinus communis] |
| 9 |
Hb_002232_400 |
0.0991233006 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH25-like [Jatropha curcas] |
| 10 |
Hb_000120_710 |
0.099287248 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 11 |
Hb_000617_340 |
0.1051449031 |
- |
- |
respiratory burst oxidase B [Manihot esculenta] |
| 12 |
Hb_098795_010 |
0.1055331495 |
- |
- |
PREDICTED: naringenin,2-oxoglutarate 3-dioxygenase-like [Jatropha curcas] |
| 13 |
Hb_006006_070 |
0.1061411981 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 14 |
Hb_003253_020 |
0.1091415539 |
transcription factor |
TF Family: B3 |
transcription factor, putative [Ricinus communis] |
| 15 |
Hb_007483_030 |
0.1095558911 |
- |
- |
- |
| 16 |
Hb_000441_170 |
0.1114911234 |
- |
- |
PREDICTED: uncharacterized protein LOC105636851 [Jatropha curcas] |
| 17 |
Hb_000948_070 |
0.112255577 |
- |
- |
hypothetical protein POPTR_0016s01720g [Populus trichocarpa] |
| 18 |
Hb_002232_370 |
0.1125445405 |
- |
- |
PREDICTED: receptor-like protein kinase HAIKU2 [Jatropha curcas] |
| 19 |
Hb_058526_010 |
0.1137883276 |
- |
- |
- |
| 20 |
Hb_003166_040 |
0.1146969776 |
- |
- |
PREDICTED: probable indole-3-acetic acid-amido synthetase GH3.1 [Jatropha curcas] |