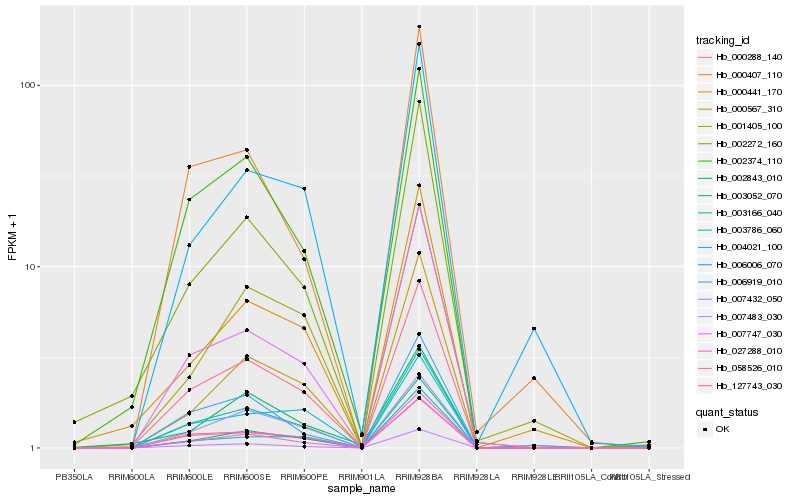
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_127743_030 |
0.0 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 2 |
Hb_003166_040 |
0.0296732016 |
- |
- |
PREDICTED: probable indole-3-acetic acid-amido synthetase GH3.1 [Jatropha curcas] |
| 3 |
Hb_058526_010 |
0.0569043694 |
- |
- |
- |
| 4 |
Hb_002374_110 |
0.0703096018 |
- |
- |
PREDICTED: uncharacterized protein LOC105638965 [Jatropha curcas] |
| 5 |
Hb_007432_050 |
0.0714356555 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB108 [Jatropha curcas] |
| 6 |
Hb_027288_010 |
0.0850796418 |
- |
- |
mutt domain protein, putative [Ricinus communis] |
| 7 |
Hb_007483_030 |
0.093002929 |
- |
- |
- |
| 8 |
Hb_007747_030 |
0.1005042387 |
- |
- |
PREDICTED: WD repeat-containing protein LWD2-like [Jatropha curcas] |
| 9 |
Hb_004021_100 |
0.1049676323 |
- |
- |
- |
| 10 |
Hb_002272_160 |
0.1051946282 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 11 |
Hb_000288_140 |
0.11668606 |
- |
- |
PREDICTED: putative cysteine-rich receptor-like protein kinase 35 [Jatropha curcas] |
| 12 |
Hb_000567_310 |
0.1171189086 |
- |
- |
Miraculin precursor, putative [Ricinus communis] |
| 13 |
Hb_003052_070 |
0.1173842499 |
- |
- |
PREDICTED: peroxidase 11 [Jatropha curcas] |
| 14 |
Hb_006919_010 |
0.117406591 |
- |
- |
PREDICTED: putative wall-associated receptor kinase-like 16 [Vitis vinifera] |
| 15 |
Hb_000441_170 |
0.120621406 |
- |
- |
PREDICTED: uncharacterized protein LOC105636851 [Jatropha curcas] |
| 16 |
Hb_003786_060 |
0.1236961646 |
- |
- |
PREDICTED: transmembrane protein 220 [Jatropha curcas] |
| 17 |
Hb_000407_110 |
0.1276307806 |
- |
- |
ripening-induced ACC oxidase [Carica papaya] |
| 18 |
Hb_006006_070 |
0.1308264917 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 19 |
Hb_002843_010 |
0.1312334715 |
- |
- |
hypothetical protein EUGRSUZ_H04984, partial [Eucalyptus grandis] |
| 20 |
Hb_001405_100 |
0.1347081995 |
- |
- |
PREDICTED: lysM domain receptor-like kinase 4 [Jatropha curcas] |