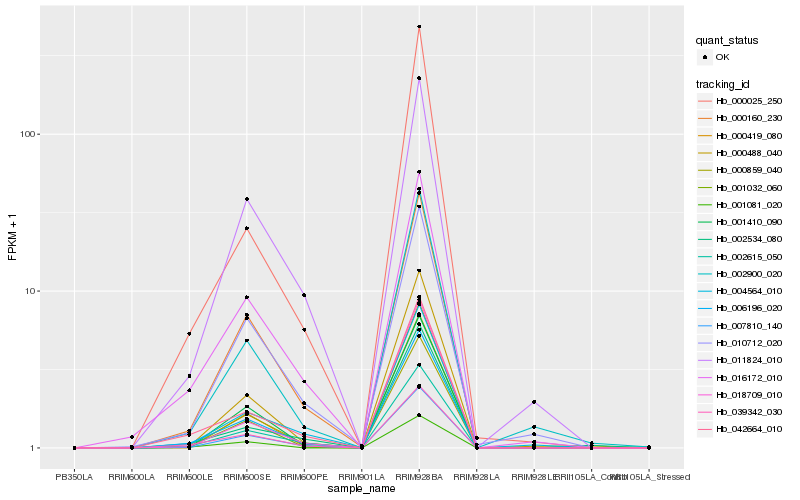
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006196_020 |
0.0 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 100-like [Populus euphratica] |
| 2 |
Hb_000160_230 |
0.0520949963 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 3 |
Hb_007810_140 |
0.0537585713 |
- |
- |
Ankyrin repeat-containing protein [Medicago truncatula] |
| 4 |
Hb_002615_050 |
0.053912388 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 5 |
Hb_042664_010 |
0.0557088182 |
- |
- |
hypothetical protein POPTR_0001s07640g, partial [Populus trichocarpa] |
| 6 |
Hb_039342_030 |
0.0563744188 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000859_040 |
0.0639357672 |
- |
- |
Early nodulin 16 precursor, putative [Ricinus communis] |
| 8 |
Hb_016172_010 |
0.0725969952 |
- |
- |
hypothetical protein JCGZ_23602 [Jatropha curcas] |
| 9 |
Hb_000488_040 |
0.0823094828 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 10 |
Hb_004564_010 |
0.0833357464 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 11 |
Hb_001032_060 |
0.0843721759 |
- |
- |
PREDICTED: ankyrin-2-like [Citrus sinensis] |
| 12 |
Hb_002900_020 |
0.0852301745 |
- |
- |
hypothetical protein POPTR_0008s06270g [Populus trichocarpa] |
| 13 |
Hb_001410_090 |
0.0865763275 |
- |
- |
transferase, putative [Ricinus communis] |
| 14 |
Hb_001081_020 |
0.0875004188 |
- |
- |
Cytochrome P450 76C2 [Triticum urartu] |
| 15 |
Hb_018709_010 |
0.0911432954 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 16 |
Hb_002534_080 |
0.0920655011 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 17 |
Hb_000025_250 |
0.0922144129 |
- |
- |
PREDICTED: flavonoid 3',5'-hydroxylase 2-like [Jatropha curcas] |
| 18 |
Hb_011824_010 |
0.0925418304 |
- |
- |
Metalloendoproteinase 1 precursor, putative [Ricinus communis] |
| 19 |
Hb_000419_080 |
0.0967995495 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-4 [Jatropha curcas] |
| 20 |
Hb_010712_020 |
0.0969788438 |
- |
- |
PREDICTED: caffeic acid 3-O-methyltransferase-like [Jatropha curcas] |