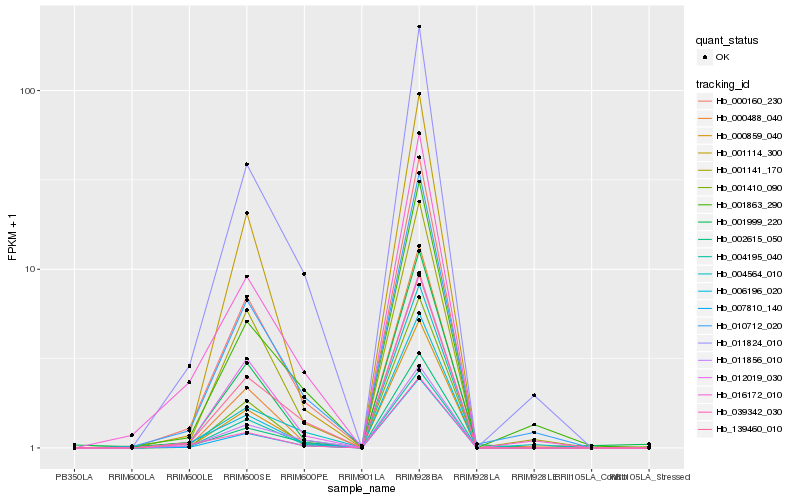
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000160_230 |
0.0 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 2 |
Hb_007810_140 |
0.014695522 |
- |
- |
Ankyrin repeat-containing protein [Medicago truncatula] |
| 3 |
Hb_000859_040 |
0.0275542582 |
- |
- |
Early nodulin 16 precursor, putative [Ricinus communis] |
| 4 |
Hb_002615_050 |
0.0415001613 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 5 |
Hb_006196_020 |
0.0520949963 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 100-like [Populus euphratica] |
| 6 |
Hb_011824_010 |
0.0613778949 |
- |
- |
Metalloendoproteinase 1 precursor, putative [Ricinus communis] |
| 7 |
Hb_010712_020 |
0.0665324294 |
- |
- |
PREDICTED: caffeic acid 3-O-methyltransferase-like [Jatropha curcas] |
| 8 |
Hb_139460_010 |
0.0677216232 |
- |
- |
PREDICTED: beta-glucosidase 18-like [Jatropha curcas] |
| 9 |
Hb_039342_030 |
0.072469811 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001410_090 |
0.0738096336 |
- |
- |
transferase, putative [Ricinus communis] |
| 11 |
Hb_016172_010 |
0.0744302923 |
- |
- |
hypothetical protein JCGZ_23602 [Jatropha curcas] |
| 12 |
Hb_001114_300 |
0.0773306186 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 13 |
Hb_001141_170 |
0.0773841641 |
- |
- |
PREDICTED: non-specific lipid-transfer protein-like protein At2g13820 [Jatropha curcas] |
| 14 |
Hb_004564_010 |
0.0823327724 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 15 |
Hb_011856_010 |
0.0831264032 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 16 |
Hb_012019_030 |
0.0850476171 |
- |
- |
hypothetical protein POPTR_0005s23360g [Populus trichocarpa] |
| 17 |
Hb_001863_290 |
0.0873497194 |
- |
- |
PREDICTED: 7-deoxyloganetin glucosyltransferase-like [Jatropha curcas] |
| 18 |
Hb_004195_040 |
0.0905206367 |
- |
- |
salicylic acid methyl transferase [Capsicum annuum] |
| 19 |
Hb_001999_220 |
0.0914677873 |
- |
- |
PREDICTED: cytochrome P450 86A1 [Jatropha curcas] |
| 20 |
Hb_000488_040 |
0.0920184808 |
- |
- |
unnamed protein product [Vitis vinifera] |