| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
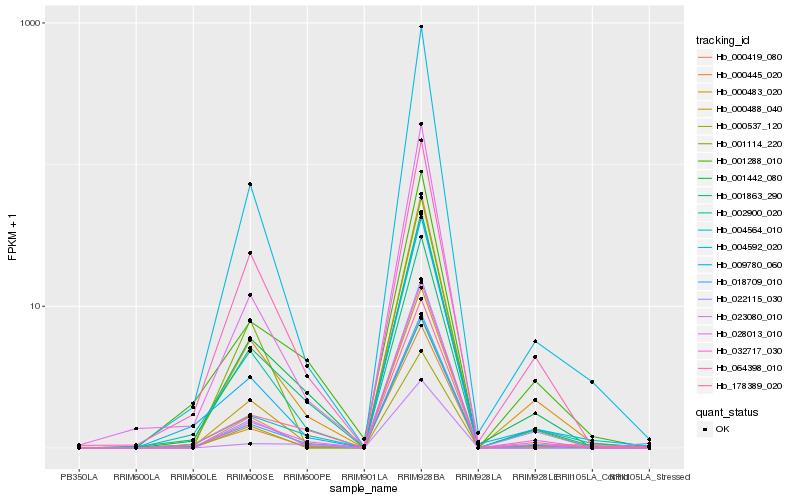
Hb_000483_020 |
0.0 |
- |
- |
PREDICTED: 7-deoxyloganetic acid glucosyltransferase-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_002900_020 |
0.0554841175 |
- |
- |
hypothetical protein POPTR_0008s06270g [Populus trichocarpa] |
| 3 |
Hb_004592_020 |
0.0688845301 |
- |
- |
PREDICTED: non-specific lipid-transfer protein 1-like [Jatropha curcas] |
| 4 |
Hb_000488_040 |
0.0801536558 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 5 |
Hb_001442_080 |
0.0804657989 |
transcription factor |
TF Family: MYB-related |
transcription factor MYB251 [Fagus crenata] |
| 6 |
Hb_032717_030 |
0.0804789586 |
- |
- |
PREDICTED: CASP-like protein 1B2 [Jatropha curcas] |
| 7 |
Hb_178389_020 |
0.0886790584 |
- |
- |
PREDICTED: cytochrome P450 86B1-like [Jatropha curcas] |
| 8 |
Hb_009780_060 |
0.090573685 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranyl geranyl pyrophosphate synthase, putative [Ricinus communis] |
| 9 |
Hb_001288_010 |
0.0934472518 |
- |
- |
- |
| 10 |
Hb_004564_010 |
0.0937786587 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 11 |
Hb_022115_030 |
0.0957807704 |
- |
- |
PREDICTED: umecyanin-like [Jatropha curcas] |
| 12 |
Hb_064398_010 |
0.0959058879 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 13 |
Hb_000537_120 |
0.0980442161 |
- |
- |
Ent-kaurene synthase B, chloroplast precursor, putative [Ricinus communis] |
| 14 |
Hb_023080_010 |
0.1016339118 |
- |
- |
PREDICTED: probable serine incorporator [Jatropha curcas] |
| 15 |
Hb_000445_020 |
0.1018570039 |
- |
- |
GABA-specific permease, putative [Ricinus communis] |
| 16 |
Hb_018709_010 |
0.1018792494 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 17 |
Hb_001863_290 |
0.1035638735 |
- |
- |
PREDICTED: 7-deoxyloganetin glucosyltransferase-like [Jatropha curcas] |
| 18 |
Hb_001114_220 |
0.1064234214 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 19 |
Hb_000419_080 |
0.106987985 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-4 [Jatropha curcas] |
| 20 |
Hb_028013_010 |
0.1081780357 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |