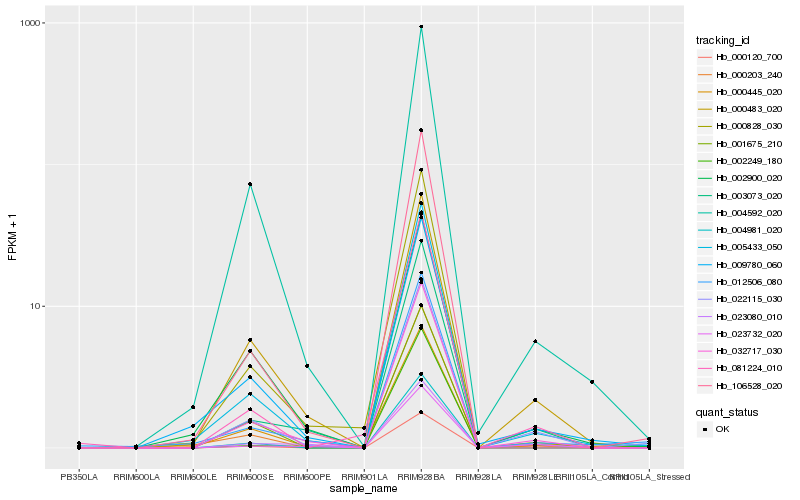
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_022115_030 |
0.0 |
- |
- |
PREDICTED: umecyanin-like [Jatropha curcas] |
| 2 |
Hb_032717_030 |
0.0692946489 |
- |
- |
PREDICTED: CASP-like protein 1B2 [Jatropha curcas] |
| 3 |
Hb_000120_700 |
0.0784435235 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 4 |
Hb_005433_050 |
0.0848544923 |
- |
- |
PREDICTED: uncharacterized protein At1g66480-like [Populus euphratica] |
| 5 |
Hb_003073_020 |
0.0856630593 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 6 |
Hb_023732_020 |
0.0954195937 |
- |
- |
PREDICTED: fatty acyl-CoA reductase 3-like [Jatropha curcas] |
| 7 |
Hb_000483_020 |
0.0957807704 |
- |
- |
PREDICTED: 7-deoxyloganetic acid glucosyltransferase-like isoform X2 [Jatropha curcas] |
| 8 |
Hb_002249_180 |
0.1010251998 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 9 |
Hb_000203_240 |
0.1027871712 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 10 |
Hb_009780_060 |
0.1036685473 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranyl geranyl pyrophosphate synthase, putative [Ricinus communis] |
| 11 |
Hb_012506_080 |
0.1044721492 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 12 |
Hb_004592_020 |
0.1050470864 |
- |
- |
PREDICTED: non-specific lipid-transfer protein 1-like [Jatropha curcas] |
| 13 |
Hb_081224_010 |
0.105786297 |
- |
- |
hypothetical protein OsJ_17572 [Oryza sativa Japonica Group] |
| 14 |
Hb_000445_020 |
0.1066010173 |
- |
- |
GABA-specific permease, putative [Ricinus communis] |
| 15 |
Hb_023080_010 |
0.1092885547 |
- |
- |
PREDICTED: probable serine incorporator [Jatropha curcas] |
| 16 |
Hb_001675_210 |
0.1118750865 |
- |
- |
PREDICTED: protein PLANT CADMIUM RESISTANCE 8-like [Jatropha curcas] |
| 17 |
Hb_000828_030 |
0.1146838233 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 18 |
Hb_106528_020 |
0.1149183166 |
- |
- |
Cationic peroxidase 2 precursor [Theobroma cacao] |
| 19 |
Hb_002900_020 |
0.11681339 |
- |
- |
hypothetical protein POPTR_0008s06270g [Populus trichocarpa] |
| 20 |
Hb_004981_020 |
0.1178093961 |
- |
- |
PREDICTED: uncharacterized protein LOC103706119 [Phoenix dactylifera] |