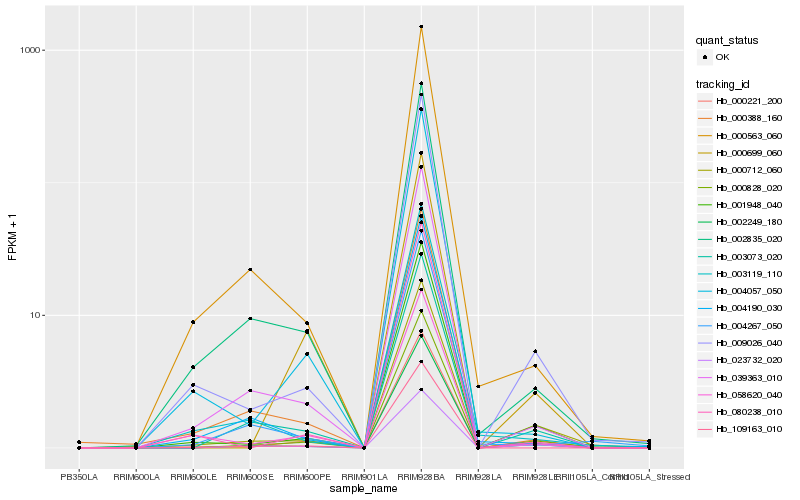
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000828_020 |
0.0 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 2 |
Hb_000221_200 |
0.0493095927 |
- |
- |
hypothetical protein JCGZ_15819 [Jatropha curcas] |
| 3 |
Hb_001948_040 |
0.0516963798 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranyl geranyl pyrophosphate synthase, putative [Ricinus communis] |
| 4 |
Hb_000712_060 |
0.053303128 |
- |
- |
PREDICTED: uncharacterized protein LOC105631517 [Jatropha curcas] |
| 5 |
Hb_009026_040 |
0.0585246763 |
- |
- |
pathogenesis-related family protein [Populus trichocarpa] |
| 6 |
Hb_003073_020 |
0.0693123636 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 7 |
Hb_002249_180 |
0.0696038543 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 8 |
Hb_039363_010 |
0.0712908153 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 9 |
Hb_002835_020 |
0.0803716684 |
- |
- |
PREDICTED: cytochrome P450 71A1-like [Jatropha curcas] |
| 10 |
Hb_023732_020 |
0.0845765155 |
- |
- |
PREDICTED: fatty acyl-CoA reductase 3-like [Jatropha curcas] |
| 11 |
Hb_004267_050 |
0.0863898207 |
- |
- |
hypothetical protein JCGZ_04357 [Jatropha curcas] |
| 12 |
Hb_004057_050 |
0.0896056091 |
- |
- |
PREDICTED: cytochrome P450 76A1-like [Jatropha curcas] |
| 13 |
Hb_000388_160 |
0.0900325994 |
- |
- |
xyloglucan endo-1 family protein [Populus trichocarpa] |
| 14 |
Hb_109163_010 |
0.0904405649 |
- |
- |
polyphenol oxidase [Hevea brasiliensis] |
| 15 |
Hb_000563_060 |
0.0906516132 |
- |
- |
PREDICTED: uncharacterized protein LOC100306381 isoform X1 [Glycine max] |
| 16 |
Hb_080238_010 |
0.091350692 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 17 |
Hb_004190_030 |
0.0957859496 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_058620_040 |
0.0961304572 |
- |
- |
Myrcene synthase, chloroplastic, putative isoform 1 [Theobroma cacao] |
| 19 |
Hb_003119_110 |
0.0972790504 |
- |
- |
PREDICTED: feruloyl CoA ortho-hydroxylase 2-like [Jatropha curcas] |
| 20 |
Hb_000699_060 |
0.0991860023 |
- |
- |
hypothetical protein POPTR_0019s03180g [Populus trichocarpa] |