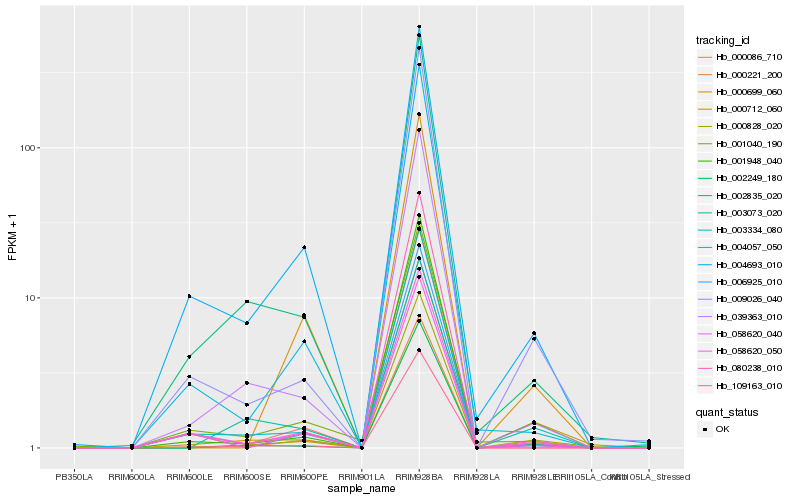
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000221_200 |
0.0 |
- |
- |
hypothetical protein JCGZ_15819 [Jatropha curcas] |
| 2 |
Hb_000828_020 |
0.0493095927 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 3 |
Hb_001948_040 |
0.0550004529 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranyl geranyl pyrophosphate synthase, putative [Ricinus communis] |
| 4 |
Hb_009026_040 |
0.0662245962 |
- |
- |
pathogenesis-related family protein [Populus trichocarpa] |
| 5 |
Hb_000699_060 |
0.0732744956 |
- |
- |
hypothetical protein POPTR_0019s03180g [Populus trichocarpa] |
| 6 |
Hb_058620_040 |
0.073463157 |
- |
- |
Myrcene synthase, chloroplastic, putative isoform 1 [Theobroma cacao] |
| 7 |
Hb_039363_010 |
0.075971131 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 8 |
Hb_000712_060 |
0.0765576603 |
- |
- |
PREDICTED: uncharacterized protein LOC105631517 [Jatropha curcas] |
| 9 |
Hb_004057_050 |
0.0772783802 |
- |
- |
PREDICTED: cytochrome P450 76A1-like [Jatropha curcas] |
| 10 |
Hb_004693_010 |
0.0784541341 |
- |
- |
hypothetical protein JCGZ_02321 [Jatropha curcas] |
| 11 |
Hb_002835_020 |
0.0820789855 |
- |
- |
PREDICTED: cytochrome P450 71A1-like [Jatropha curcas] |
| 12 |
Hb_003073_020 |
0.0838344529 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 13 |
Hb_058620_050 |
0.0852829063 |
- |
- |
PREDICTED: terpene synthase 10-like [Jatropha curcas] |
| 14 |
Hb_080238_010 |
0.0857416774 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 15 |
Hb_003334_080 |
0.0869477968 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 16 |
Hb_006925_010 |
0.0873601089 |
- |
- |
PREDICTED: uncharacterized protein LOC101777526 [Setaria italica] |
| 17 |
Hb_000086_710 |
0.087673311 |
- |
- |
PREDICTED: feruloyl CoA ortho-hydroxylase 2-like [Jatropha curcas] |
| 18 |
Hb_002249_180 |
0.0884702362 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 19 |
Hb_001040_190 |
0.088710536 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor B-2a-like [Jatropha curcas] |
| 20 |
Hb_109163_010 |
0.0906338867 |
- |
- |
polyphenol oxidase [Hevea brasiliensis] |