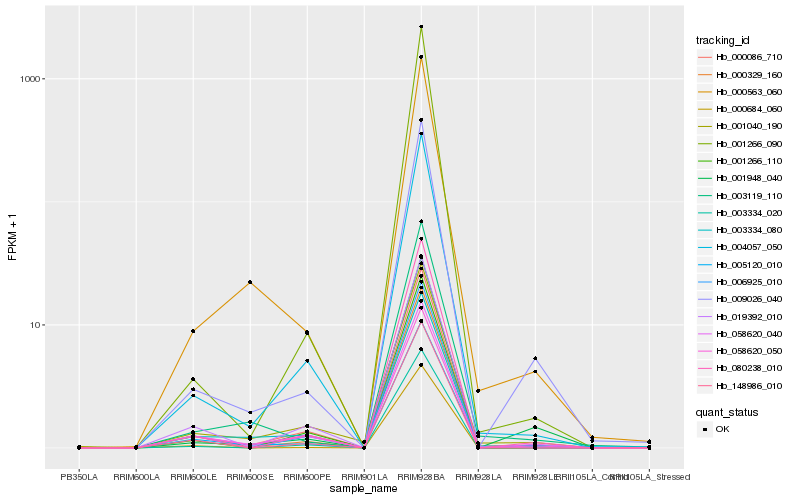
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000329_160 |
0.0 |
- |
- |
PREDICTED: pectinesterase 2 isoform X1 [Jatropha curcas] |
| 2 |
Hb_006925_010 |
0.0435802844 |
- |
- |
PREDICTED: uncharacterized protein LOC101777526 [Setaria italica] |
| 3 |
Hb_004057_050 |
0.0493320612 |
- |
- |
PREDICTED: cytochrome P450 76A1-like [Jatropha curcas] |
| 4 |
Hb_019392_010 |
0.0503035647 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable S-adenosylmethionine-dependent methyltransferase At5g38100 [Jatropha curcas] |
| 5 |
Hb_148986_010 |
0.0522159394 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_080238_010 |
0.0532478056 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 7 |
Hb_000086_710 |
0.0576450623 |
- |
- |
PREDICTED: feruloyl CoA ortho-hydroxylase 2-like [Jatropha curcas] |
| 8 |
Hb_000684_060 |
0.0583807037 |
- |
- |
hypothetical protein POPTR_0006s13170g [Populus trichocarpa] |
| 9 |
Hb_005120_010 |
0.0622025407 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 10 |
Hb_058620_040 |
0.062298141 |
- |
- |
Myrcene synthase, chloroplastic, putative isoform 1 [Theobroma cacao] |
| 11 |
Hb_003334_020 |
0.0642909919 |
- |
- |
hypothetical protein CISIN_1g041546mg [Citrus sinensis] |
| 12 |
Hb_001266_110 |
0.0661919985 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_058620_050 |
0.0704597021 |
- |
- |
PREDICTED: terpene synthase 10-like [Jatropha curcas] |
| 14 |
Hb_009026_040 |
0.070800889 |
- |
- |
pathogenesis-related family protein [Populus trichocarpa] |
| 15 |
Hb_001040_190 |
0.0746359893 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor B-2a-like [Jatropha curcas] |
| 16 |
Hb_003119_110 |
0.0763671674 |
- |
- |
PREDICTED: feruloyl CoA ortho-hydroxylase 2-like [Jatropha curcas] |
| 17 |
Hb_000563_060 |
0.0796121488 |
- |
- |
PREDICTED: uncharacterized protein LOC100306381 isoform X1 [Glycine max] |
| 18 |
Hb_003334_080 |
0.0797697338 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 19 |
Hb_001266_090 |
0.0812518351 |
- |
- |
- |
| 20 |
Hb_001948_040 |
0.0832064975 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranyl geranyl pyrophosphate synthase, putative [Ricinus communis] |