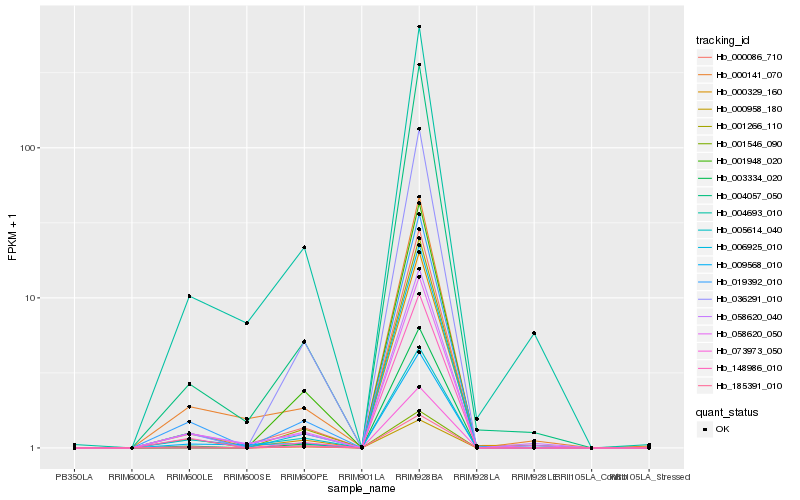
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000958_180 |
0.0 |
- |
- |
hypothetical protein CISIN_1g043061mg, partial [Citrus sinensis] |
| 2 |
Hb_185391_010 |
0.0358540837 |
- |
- |
PREDICTED: metalloendoproteinase 1-like [Populus euphratica] |
| 3 |
Hb_019392_010 |
0.0495682202 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable S-adenosylmethionine-dependent methyltransferase At5g38100 [Jatropha curcas] |
| 4 |
Hb_001266_110 |
0.0602060593 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_006925_010 |
0.0647713847 |
- |
- |
PREDICTED: uncharacterized protein LOC101777526 [Setaria italica] |
| 6 |
Hb_000086_710 |
0.0648480416 |
- |
- |
PREDICTED: feruloyl CoA ortho-hydroxylase 2-like [Jatropha curcas] |
| 7 |
Hb_058620_050 |
0.0651039839 |
- |
- |
PREDICTED: terpene synthase 10-like [Jatropha curcas] |
| 8 |
Hb_036291_010 |
0.0660755177 |
- |
- |
PREDICTED: alpha carbonic anhydrase 7-like [Jatropha curcas] |
| 9 |
Hb_148986_010 |
0.0661846533 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 10 |
Hb_003334_020 |
0.0703414749 |
- |
- |
hypothetical protein CISIN_1g041546mg [Citrus sinensis] |
| 11 |
Hb_058620_040 |
0.0781257534 |
- |
- |
Myrcene synthase, chloroplastic, putative isoform 1 [Theobroma cacao] |
| 12 |
Hb_004057_050 |
0.0818515013 |
- |
- |
PREDICTED: cytochrome P450 76A1-like [Jatropha curcas] |
| 13 |
Hb_009568_010 |
0.0869579204 |
- |
- |
hypothetical protein POPTR_0001s46700g [Populus trichocarpa] |
| 14 |
Hb_001948_020 |
0.0873189847 |
- |
- |
- |
| 15 |
Hb_001546_090 |
0.0907233954 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB114-like [Jatropha curcas] |
| 16 |
Hb_000329_160 |
0.0911259337 |
- |
- |
PREDICTED: pectinesterase 2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_004693_010 |
0.0912449719 |
- |
- |
hypothetical protein JCGZ_02321 [Jatropha curcas] |
| 18 |
Hb_005614_040 |
0.0913365282 |
- |
- |
unnamed protein product [Coffea canephora] |
| 19 |
Hb_073973_050 |
0.0917317656 |
transcription factor |
TF Family: GRAS |
PREDICTED: nodulation-signaling pathway 2 protein [Jatropha curcas] |
| 20 |
Hb_000141_070 |
0.0929809719 |
- |
- |
l-lactate dehydrogenase, putative [Ricinus communis] |