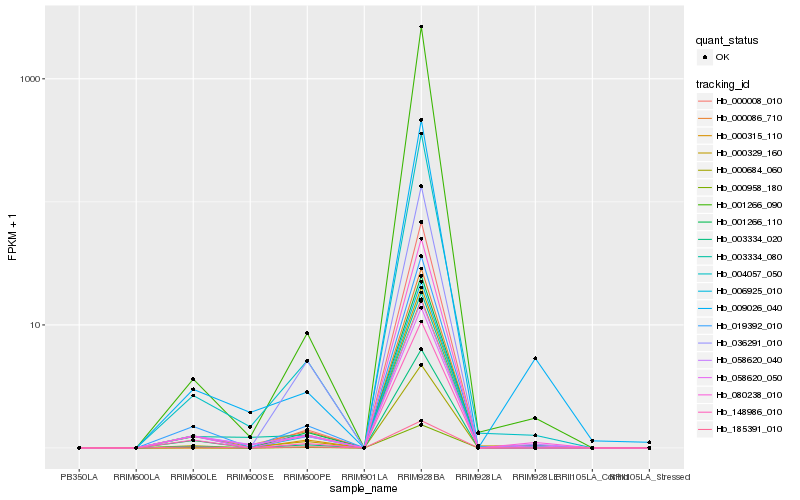
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006925_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC101777526 [Setaria italica] |
| 2 |
Hb_148986_010 |
0.0343423158 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 3 |
Hb_019392_010 |
0.0390393814 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable S-adenosylmethionine-dependent methyltransferase At5g38100 [Jatropha curcas] |
| 4 |
Hb_001266_110 |
0.039548658 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_003334_020 |
0.040917743 |
- |
- |
hypothetical protein CISIN_1g041546mg [Citrus sinensis] |
| 6 |
Hb_080238_010 |
0.0417531835 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 7 |
Hb_000329_160 |
0.0435802844 |
- |
- |
PREDICTED: pectinesterase 2 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000086_710 |
0.0456826752 |
- |
- |
PREDICTED: feruloyl CoA ortho-hydroxylase 2-like [Jatropha curcas] |
| 9 |
Hb_004057_050 |
0.0487223681 |
- |
- |
PREDICTED: cytochrome P450 76A1-like [Jatropha curcas] |
| 10 |
Hb_000684_060 |
0.0564912433 |
- |
- |
hypothetical protein POPTR_0006s13170g [Populus trichocarpa] |
| 11 |
Hb_058620_040 |
0.0589836638 |
- |
- |
Myrcene synthase, chloroplastic, putative isoform 1 [Theobroma cacao] |
| 12 |
Hb_058620_050 |
0.0627414053 |
- |
- |
PREDICTED: terpene synthase 10-like [Jatropha curcas] |
| 13 |
Hb_000958_180 |
0.0647713847 |
- |
- |
hypothetical protein CISIN_1g043061mg, partial [Citrus sinensis] |
| 14 |
Hb_009026_040 |
0.076901823 |
- |
- |
pathogenesis-related family protein [Populus trichocarpa] |
| 15 |
Hb_001266_090 |
0.0787244338 |
- |
- |
- |
| 16 |
Hb_003334_080 |
0.0803646824 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 17 |
Hb_185391_010 |
0.0815832227 |
- |
- |
PREDICTED: metalloendoproteinase 1-like [Populus euphratica] |
| 18 |
Hb_000315_110 |
0.0815941898 |
- |
- |
PREDICTED: gibberellin 3-beta-dioxygenase 1-like [Jatropha curcas] |
| 19 |
Hb_036291_010 |
0.0836985381 |
- |
- |
PREDICTED: alpha carbonic anhydrase 7-like [Jatropha curcas] |
| 20 |
Hb_000008_010 |
0.0849914935 |
- |
- |
PREDICTED: alpha carbonic anhydrase 7-like [Jatropha curcas] |