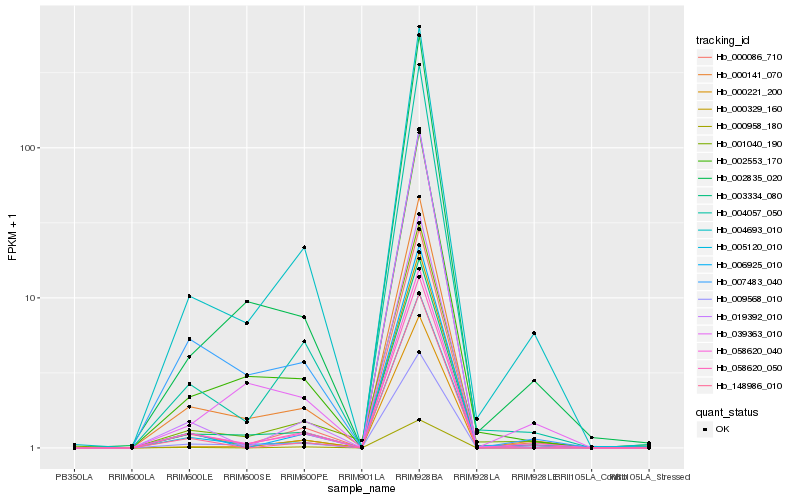
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_058620_040 |
0.0 |
- |
- |
Myrcene synthase, chloroplastic, putative isoform 1 [Theobroma cacao] |
| 2 |
Hb_058620_050 |
0.020985638 |
- |
- |
PREDICTED: terpene synthase 10-like [Jatropha curcas] |
| 3 |
Hb_003334_080 |
0.0408109122 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 4 |
Hb_000141_070 |
0.044517023 |
- |
- |
l-lactate dehydrogenase, putative [Ricinus communis] |
| 5 |
Hb_004693_010 |
0.0540060566 |
- |
- |
hypothetical protein JCGZ_02321 [Jatropha curcas] |
| 6 |
Hb_006925_010 |
0.0589836638 |
- |
- |
PREDICTED: uncharacterized protein LOC101777526 [Setaria italica] |
| 7 |
Hb_000086_710 |
0.0620428138 |
- |
- |
PREDICTED: feruloyl CoA ortho-hydroxylase 2-like [Jatropha curcas] |
| 8 |
Hb_000329_160 |
0.062298141 |
- |
- |
PREDICTED: pectinesterase 2 isoform X1 [Jatropha curcas] |
| 9 |
Hb_009568_010 |
0.0670164271 |
- |
- |
hypothetical protein POPTR_0001s46700g [Populus trichocarpa] |
| 10 |
Hb_001040_190 |
0.0676807524 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor B-2a-like [Jatropha curcas] |
| 11 |
Hb_019392_010 |
0.0678984033 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable S-adenosylmethionine-dependent methyltransferase At5g38100 [Jatropha curcas] |
| 12 |
Hb_005120_010 |
0.068380806 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 13 |
Hb_007483_040 |
0.0688931701 |
- |
- |
PREDICTED: protein SRG1-like [Jatropha curcas] |
| 14 |
Hb_002835_020 |
0.071244858 |
- |
- |
PREDICTED: cytochrome P450 71A1-like [Jatropha curcas] |
| 15 |
Hb_004057_050 |
0.0718565382 |
- |
- |
PREDICTED: cytochrome P450 76A1-like [Jatropha curcas] |
| 16 |
Hb_000221_200 |
0.073463157 |
- |
- |
hypothetical protein JCGZ_15819 [Jatropha curcas] |
| 17 |
Hb_002553_170 |
0.0736817228 |
- |
- |
PREDICTED: early nodulin-75-like [Jatropha curcas] |
| 18 |
Hb_148986_010 |
0.0738678124 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 19 |
Hb_039363_010 |
0.0772800072 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 20 |
Hb_000958_180 |
0.0781257534 |
- |
- |
hypothetical protein CISIN_1g043061mg, partial [Citrus sinensis] |