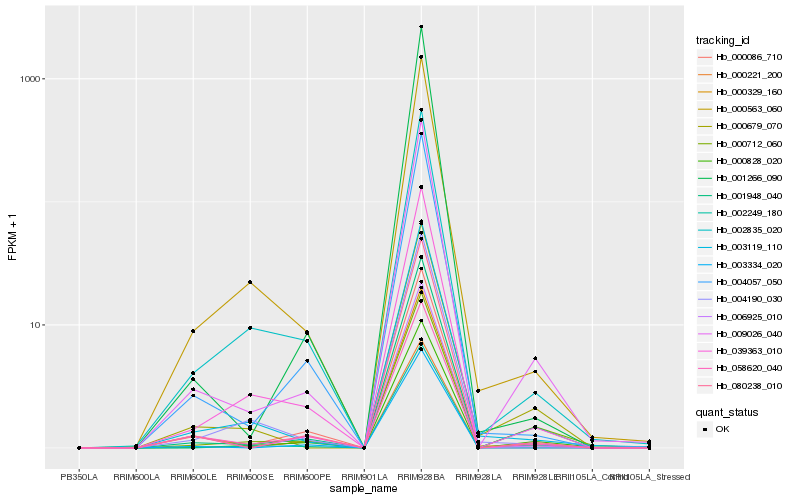
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009026_040 |
0.0 |
- |
- |
pathogenesis-related family protein [Populus trichocarpa] |
| 2 |
Hb_001948_040 |
0.0265225514 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranyl geranyl pyrophosphate synthase, putative [Ricinus communis] |
| 3 |
Hb_080238_010 |
0.0554124844 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 4 |
Hb_000828_020 |
0.0585246763 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 5 |
Hb_000221_200 |
0.0662245962 |
- |
- |
hypothetical protein JCGZ_15819 [Jatropha curcas] |
| 6 |
Hb_002249_180 |
0.070553383 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 7 |
Hb_000329_160 |
0.070800889 |
- |
- |
PREDICTED: pectinesterase 2 isoform X1 [Jatropha curcas] |
| 8 |
Hb_039363_010 |
0.0714815288 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 9 |
Hb_004057_050 |
0.0721448849 |
- |
- |
PREDICTED: cytochrome P450 76A1-like [Jatropha curcas] |
| 10 |
Hb_003119_110 |
0.0744684707 |
- |
- |
PREDICTED: feruloyl CoA ortho-hydroxylase 2-like [Jatropha curcas] |
| 11 |
Hb_000712_060 |
0.0760284072 |
- |
- |
PREDICTED: uncharacterized protein LOC105631517 [Jatropha curcas] |
| 12 |
Hb_001266_090 |
0.0764603634 |
- |
- |
- |
| 13 |
Hb_006925_010 |
0.076901823 |
- |
- |
PREDICTED: uncharacterized protein LOC101777526 [Setaria italica] |
| 14 |
Hb_000679_070 |
0.0772340008 |
- |
- |
hypothetical protein POPTR_0006s26840g [Populus trichocarpa] |
| 15 |
Hb_000563_060 |
0.0778505829 |
- |
- |
PREDICTED: uncharacterized protein LOC100306381 isoform X1 [Glycine max] |
| 16 |
Hb_004190_030 |
0.0811630256 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_002835_020 |
0.0815042859 |
- |
- |
PREDICTED: cytochrome P450 71A1-like [Jatropha curcas] |
| 18 |
Hb_058620_040 |
0.0821234218 |
- |
- |
Myrcene synthase, chloroplastic, putative isoform 1 [Theobroma cacao] |
| 19 |
Hb_000086_710 |
0.0846875109 |
- |
- |
PREDICTED: feruloyl CoA ortho-hydroxylase 2-like [Jatropha curcas] |
| 20 |
Hb_003334_020 |
0.084966767 |
- |
- |
hypothetical protein CISIN_1g041546mg [Citrus sinensis] |