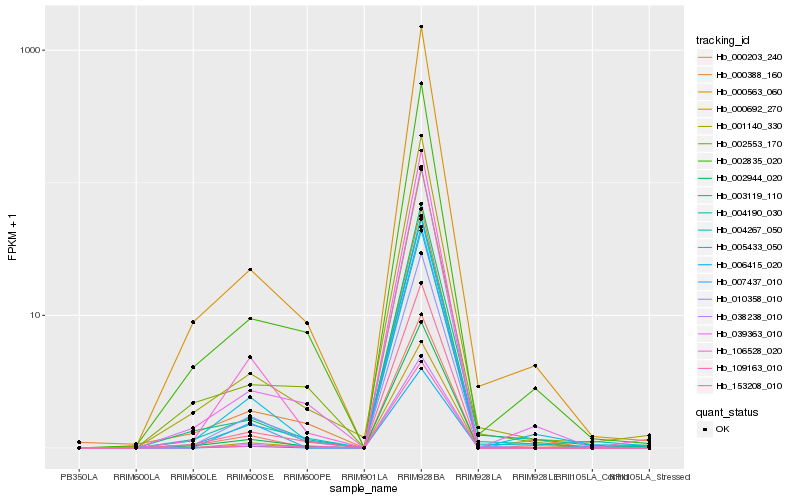
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004267_050 |
0.0 |
- |
- |
hypothetical protein JCGZ_04357 [Jatropha curcas] |
| 2 |
Hb_004190_030 |
0.037692432 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 3 |
Hb_001140_330 |
0.0399800157 |
- |
- |
PREDICTED: uncharacterized protein LOC105142675 [Populus euphratica] |
| 4 |
Hb_003119_110 |
0.0485062857 |
- |
- |
PREDICTED: feruloyl CoA ortho-hydroxylase 2-like [Jatropha curcas] |
| 5 |
Hb_000563_060 |
0.0489289709 |
- |
- |
PREDICTED: uncharacterized protein LOC100306381 isoform X1 [Glycine max] |
| 6 |
Hb_002944_020 |
0.0571313119 |
transcription factor |
TF Family: bZIP |
hypothetical protein POPTR_0006s05770g, partial [Populus trichocarpa] |
| 7 |
Hb_010358_010 |
0.0577060782 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 8 |
Hb_000692_270 |
0.0605784703 |
- |
- |
PREDICTED: ornithine decarboxylase-like [Jatropha curcas] |
| 9 |
Hb_039363_010 |
0.0621206662 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 10 |
Hb_002835_020 |
0.0622544072 |
- |
- |
PREDICTED: cytochrome P450 71A1-like [Jatropha curcas] |
| 11 |
Hb_038238_010 |
0.0650783147 |
- |
- |
hypothetical protein POPTR_0001s30980g [Populus trichocarpa] |
| 12 |
Hb_106528_020 |
0.0660873339 |
- |
- |
Cationic peroxidase 2 precursor [Theobroma cacao] |
| 13 |
Hb_109163_010 |
0.066299598 |
- |
- |
polyphenol oxidase [Hevea brasiliensis] |
| 14 |
Hb_005433_050 |
0.0666746885 |
- |
- |
PREDICTED: uncharacterized protein At1g66480-like [Populus euphratica] |
| 15 |
Hb_153208_010 |
0.0669957569 |
- |
- |
Cationic peroxidase 2 precursor, putative [Ricinus communis] |
| 16 |
Hb_000388_160 |
0.0725748245 |
- |
- |
xyloglucan endo-1 family protein [Populus trichocarpa] |
| 17 |
Hb_007437_010 |
0.0774117859 |
- |
- |
Polygalacturonase precursor, putative [Ricinus communis] |
| 18 |
Hb_006415_020 |
0.0780668271 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 4-like [Jatropha curcas] |
| 19 |
Hb_002553_170 |
0.0790172044 |
- |
- |
PREDICTED: early nodulin-75-like [Jatropha curcas] |
| 20 |
Hb_000203_240 |
0.0805061889 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |