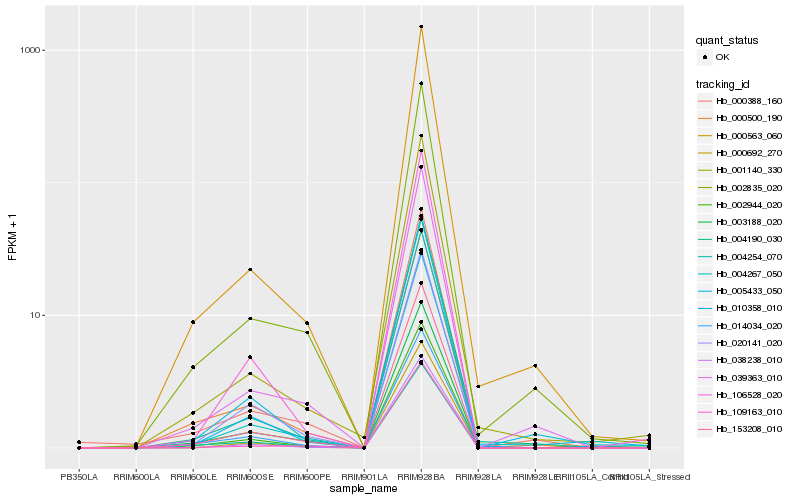
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002944_020 |
0.0 |
transcription factor |
TF Family: bZIP |
hypothetical protein POPTR_0006s05770g, partial [Populus trichocarpa] |
| 2 |
Hb_010358_010 |
0.0407479362 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 3 |
Hb_153208_010 |
0.0411426156 |
- |
- |
Cationic peroxidase 2 precursor, putative [Ricinus communis] |
| 4 |
Hb_106528_020 |
0.0554519614 |
- |
- |
Cationic peroxidase 2 precursor [Theobroma cacao] |
| 5 |
Hb_000692_270 |
0.0557708737 |
- |
- |
PREDICTED: ornithine decarboxylase-like [Jatropha curcas] |
| 6 |
Hb_003188_020 |
0.0559517248 |
- |
- |
chitinase, putative [Ricinus communis] |
| 7 |
Hb_014034_020 |
0.0561906144 |
- |
- |
hypothetical protein POPTR_0075s00200g, partial [Populus trichocarpa] |
| 8 |
Hb_000563_060 |
0.0562925746 |
- |
- |
PREDICTED: uncharacterized protein LOC100306381 isoform X1 [Glycine max] |
| 9 |
Hb_004267_050 |
0.0571313119 |
- |
- |
hypothetical protein JCGZ_04357 [Jatropha curcas] |
| 10 |
Hb_004190_030 |
0.0596173824 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 11 |
Hb_001140_330 |
0.0607837864 |
- |
- |
PREDICTED: uncharacterized protein LOC105142675 [Populus euphratica] |
| 12 |
Hb_000500_190 |
0.0611279674 |
- |
- |
hypothetical protein JCGZ_21655 [Jatropha curcas] |
| 13 |
Hb_038238_010 |
0.0612671644 |
- |
- |
hypothetical protein POPTR_0001s30980g [Populus trichocarpa] |
| 14 |
Hb_005433_050 |
0.0613972076 |
- |
- |
PREDICTED: uncharacterized protein At1g66480-like [Populus euphratica] |
| 15 |
Hb_020141_020 |
0.0627459966 |
- |
- |
- |
| 16 |
Hb_109163_010 |
0.0655528328 |
- |
- |
polyphenol oxidase [Hevea brasiliensis] |
| 17 |
Hb_002835_020 |
0.0664794951 |
- |
- |
PREDICTED: cytochrome P450 71A1-like [Jatropha curcas] |
| 18 |
Hb_039363_010 |
0.0665963523 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 19 |
Hb_004254_070 |
0.0684896113 |
transcription factor |
TF Family: NAC |
hypothetical protein POPTR_0015s03700g [Populus trichocarpa] |
| 20 |
Hb_000388_160 |
0.0692868502 |
- |
- |
xyloglucan endo-1 family protein [Populus trichocarpa] |