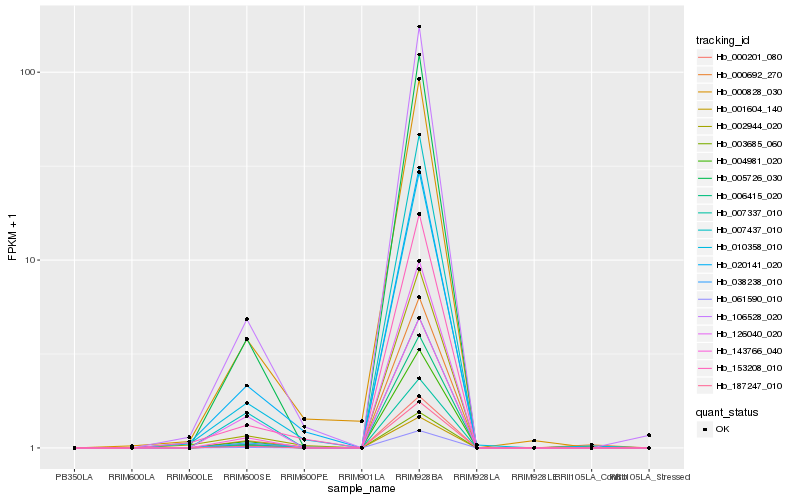
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_061590_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_23475 [Jatropha curcas] |
| 2 |
Hb_005726_030 |
0.0012890217 |
- |
- |
hypothetical protein CISIN_1g020511mg [Citrus sinensis] |
| 3 |
Hb_187247_010 |
0.0128725195 |
- |
- |
PREDICTED: cytochrome P450 86B1-like [Jatropha curcas] |
| 4 |
Hb_007337_010 |
0.0243487181 |
- |
- |
PREDICTED: putative receptor protein kinase ZmPK1 [Jatropha curcas] |
| 5 |
Hb_143766_040 |
0.0257954867 |
- |
- |
PREDICTED: probable caffeoyl-CoA O-methyltransferase At4g26220 [Jatropha curcas] |
| 6 |
Hb_004981_020 |
0.028705359 |
- |
- |
PREDICTED: uncharacterized protein LOC103706119 [Phoenix dactylifera] |
| 7 |
Hb_007437_010 |
0.036104605 |
- |
- |
Polygalacturonase precursor, putative [Ricinus communis] |
| 8 |
Hb_106528_020 |
0.0413437853 |
- |
- |
Cationic peroxidase 2 precursor [Theobroma cacao] |
| 9 |
Hb_038238_010 |
0.0422621291 |
- |
- |
hypothetical protein POPTR_0001s30980g [Populus trichocarpa] |
| 10 |
Hb_006415_020 |
0.0464293891 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 4-like [Jatropha curcas] |
| 11 |
Hb_010358_010 |
0.0613579984 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 12 |
Hb_126040_020 |
0.0636513838 |
- |
- |
- |
| 13 |
Hb_000201_080 |
0.0685683621 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 14 |
Hb_000692_270 |
0.0714772342 |
- |
- |
PREDICTED: ornithine decarboxylase-like [Jatropha curcas] |
| 15 |
Hb_153208_010 |
0.0728847491 |
- |
- |
Cationic peroxidase 2 precursor, putative [Ricinus communis] |
| 16 |
Hb_001604_140 |
0.073178336 |
- |
- |
PREDICTED: ABC transporter G family member 10-like [Populus euphratica] |
| 17 |
Hb_002944_020 |
0.0734185057 |
transcription factor |
TF Family: bZIP |
hypothetical protein POPTR_0006s05770g, partial [Populus trichocarpa] |
| 18 |
Hb_000828_030 |
0.0755468964 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 19 |
Hb_020141_020 |
0.075916002 |
- |
- |
- |
| 20 |
Hb_003685_060 |
0.075956001 |
- |
- |
PREDICTED: laccase-15 [Jatropha curcas] |