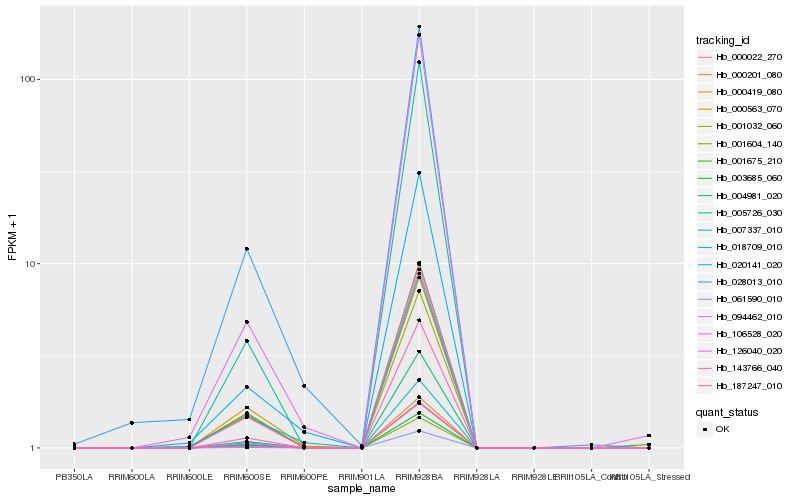
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_126040_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_000201_080 |
0.0050053096 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 3 |
Hb_001604_140 |
0.0097063353 |
- |
- |
PREDICTED: ABC transporter G family member 10-like [Populus euphratica] |
| 4 |
Hb_003685_060 |
0.0125424995 |
- |
- |
PREDICTED: laccase-15 [Jatropha curcas] |
| 5 |
Hb_000022_270 |
0.0150952943 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_004981_020 |
0.0352540973 |
- |
- |
PREDICTED: uncharacterized protein LOC103706119 [Phoenix dactylifera] |
| 7 |
Hb_143766_040 |
0.0381593368 |
- |
- |
PREDICTED: probable caffeoyl-CoA O-methyltransferase At4g26220 [Jatropha curcas] |
| 8 |
Hb_007337_010 |
0.0396017771 |
- |
- |
PREDICTED: putative receptor protein kinase ZmPK1 [Jatropha curcas] |
| 9 |
Hb_094462_010 |
0.0484032733 |
- |
- |
hypothetical protein CISIN_1g033698mg [Citrus sinensis] |
| 10 |
Hb_001675_210 |
0.0491109465 |
- |
- |
PREDICTED: protein PLANT CADMIUM RESISTANCE 8-like [Jatropha curcas] |
| 11 |
Hb_187247_010 |
0.0509928985 |
- |
- |
PREDICTED: cytochrome P450 86B1-like [Jatropha curcas] |
| 12 |
Hb_061590_010 |
0.0636513838 |
- |
- |
hypothetical protein JCGZ_23475 [Jatropha curcas] |
| 13 |
Hb_001032_060 |
0.0640472671 |
- |
- |
PREDICTED: ankyrin-2-like [Citrus sinensis] |
| 14 |
Hb_005726_030 |
0.0649113684 |
- |
- |
hypothetical protein CISIN_1g020511mg [Citrus sinensis] |
| 15 |
Hb_000419_080 |
0.0655756693 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-4 [Jatropha curcas] |
| 16 |
Hb_028013_010 |
0.0717044277 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 17 |
Hb_020141_020 |
0.0731600673 |
- |
- |
- |
| 18 |
Hb_018709_010 |
0.0731954505 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 19 |
Hb_000563_070 |
0.0754538011 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase LOG2 [Sesamum indicum] |
| 20 |
Hb_106528_020 |
0.0784333729 |
- |
- |
Cationic peroxidase 2 precursor [Theobroma cacao] |