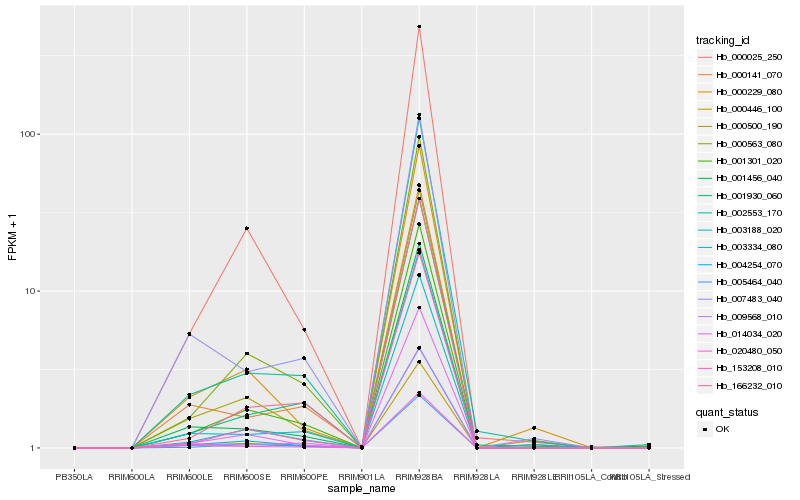
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000446_100 |
0.0 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 2 |
Hb_004254_070 |
0.0202221851 |
transcription factor |
TF Family: NAC |
hypothetical protein POPTR_0015s03700g [Populus trichocarpa] |
| 3 |
Hb_001301_020 |
0.0290336909 |
- |
- |
hypothetical protein JCGZ_06575 [Jatropha curcas] |
| 4 |
Hb_003188_020 |
0.0331316236 |
- |
- |
chitinase, putative [Ricinus communis] |
| 5 |
Hb_000500_190 |
0.0368161977 |
- |
- |
hypothetical protein JCGZ_21655 [Jatropha curcas] |
| 6 |
Hb_014034_020 |
0.0369199378 |
- |
- |
hypothetical protein POPTR_0075s00200g, partial [Populus trichocarpa] |
| 7 |
Hb_001456_040 |
0.0424030279 |
- |
- |
NAD dependent epimerase/dehydratase, putative [Ricinus communis] |
| 8 |
Hb_000563_080 |
0.0441663281 |
- |
- |
- |
| 9 |
Hb_009568_010 |
0.0468334503 |
- |
- |
hypothetical protein POPTR_0001s46700g [Populus trichocarpa] |
| 10 |
Hb_020480_050 |
0.0486469022 |
- |
- |
putative wall-associated kinase family protein [Populus trichocarpa] |
| 11 |
Hb_005464_040 |
0.0567069074 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 12 |
Hb_000025_250 |
0.0576302176 |
- |
- |
PREDICTED: flavonoid 3',5'-hydroxylase 2-like [Jatropha curcas] |
| 13 |
Hb_002553_170 |
0.0576449932 |
- |
- |
PREDICTED: early nodulin-75-like [Jatropha curcas] |
| 14 |
Hb_003334_080 |
0.057846476 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 15 |
Hb_000229_080 |
0.0603677106 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 16 |
Hb_153208_010 |
0.0615463756 |
- |
- |
Cationic peroxidase 2 precursor, putative [Ricinus communis] |
| 17 |
Hb_000141_070 |
0.0638513243 |
- |
- |
l-lactate dehydrogenase, putative [Ricinus communis] |
| 18 |
Hb_007483_040 |
0.0646610662 |
- |
- |
PREDICTED: protein SRG1-like [Jatropha curcas] |
| 19 |
Hb_166232_010 |
0.0651053098 |
- |
- |
hypothetical protein JCGZ_22831 [Jatropha curcas] |
| 20 |
Hb_001930_060 |
0.0701743888 |
- |
- |
PREDICTED: cytochrome P450 71D9-like [Fragaria vesca subsp. vesca] |