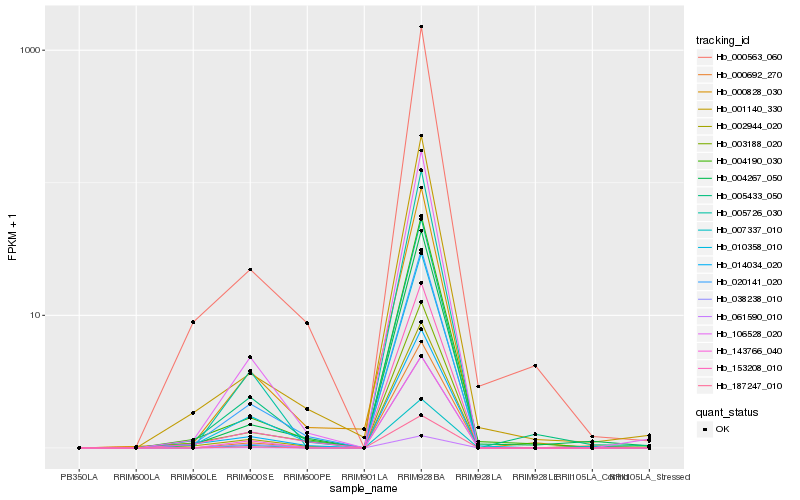
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010358_010 |
0.0 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 2 |
Hb_002944_020 |
0.0407479362 |
transcription factor |
TF Family: bZIP |
hypothetical protein POPTR_0006s05770g, partial [Populus trichocarpa] |
| 3 |
Hb_153208_010 |
0.0443247246 |
- |
- |
Cationic peroxidase 2 precursor, putative [Ricinus communis] |
| 4 |
Hb_106528_020 |
0.0448193019 |
- |
- |
Cationic peroxidase 2 precursor [Theobroma cacao] |
| 5 |
Hb_020141_020 |
0.047397904 |
- |
- |
- |
| 6 |
Hb_038238_010 |
0.0524341036 |
- |
- |
hypothetical protein POPTR_0001s30980g [Populus trichocarpa] |
| 7 |
Hb_004190_030 |
0.0530367035 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 8 |
Hb_000563_060 |
0.0566948988 |
- |
- |
PREDICTED: uncharacterized protein LOC100306381 isoform X1 [Glycine max] |
| 9 |
Hb_004267_050 |
0.0577060782 |
- |
- |
hypothetical protein JCGZ_04357 [Jatropha curcas] |
| 10 |
Hb_001140_330 |
0.0595900185 |
- |
- |
PREDICTED: uncharacterized protein LOC105142675 [Populus euphratica] |
| 11 |
Hb_003188_020 |
0.0601810541 |
- |
- |
chitinase, putative [Ricinus communis] |
| 12 |
Hb_005433_050 |
0.0605530821 |
- |
- |
PREDICTED: uncharacterized protein At1g66480-like [Populus euphratica] |
| 13 |
Hb_187247_010 |
0.0612484594 |
- |
- |
PREDICTED: cytochrome P450 86B1-like [Jatropha curcas] |
| 14 |
Hb_014034_020 |
0.0612679457 |
- |
- |
hypothetical protein POPTR_0075s00200g, partial [Populus trichocarpa] |
| 15 |
Hb_061590_010 |
0.0613579984 |
- |
- |
hypothetical protein JCGZ_23475 [Jatropha curcas] |
| 16 |
Hb_005726_030 |
0.061516928 |
- |
- |
hypothetical protein CISIN_1g020511mg [Citrus sinensis] |
| 17 |
Hb_000828_030 |
0.0631303299 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 18 |
Hb_007337_010 |
0.0633902572 |
- |
- |
PREDICTED: putative receptor protein kinase ZmPK1 [Jatropha curcas] |
| 19 |
Hb_143766_040 |
0.0638018399 |
- |
- |
PREDICTED: probable caffeoyl-CoA O-methyltransferase At4g26220 [Jatropha curcas] |
| 20 |
Hb_000692_270 |
0.064317731 |
- |
- |
PREDICTED: ornithine decarboxylase-like [Jatropha curcas] |