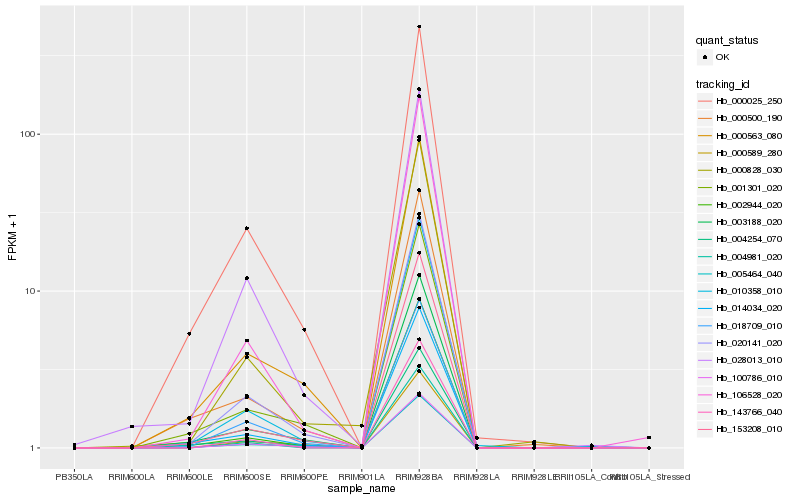
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_020141_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_018709_010 |
0.0405076497 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 3 |
Hb_000563_080 |
0.0450639156 |
- |
- |
- |
| 4 |
Hb_003188_020 |
0.0451262434 |
- |
- |
chitinase, putative [Ricinus communis] |
| 5 |
Hb_010358_010 |
0.047397904 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 6 |
Hb_153208_010 |
0.0478025829 |
- |
- |
Cationic peroxidase 2 precursor, putative [Ricinus communis] |
| 7 |
Hb_014034_020 |
0.0478552156 |
- |
- |
hypothetical protein POPTR_0075s00200g, partial [Populus trichocarpa] |
| 8 |
Hb_100786_010 |
0.0485382644 |
- |
- |
hypothetical protein B456_008G202600 [Gossypium raimondii] |
| 9 |
Hb_000025_250 |
0.0486521244 |
- |
- |
PREDICTED: flavonoid 3',5'-hydroxylase 2-like [Jatropha curcas] |
| 10 |
Hb_028013_010 |
0.0501361629 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 11 |
Hb_001301_020 |
0.0561800549 |
- |
- |
hypothetical protein JCGZ_06575 [Jatropha curcas] |
| 12 |
Hb_000828_030 |
0.0576443046 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 13 |
Hb_106528_020 |
0.0584415704 |
- |
- |
Cationic peroxidase 2 precursor [Theobroma cacao] |
| 14 |
Hb_004254_070 |
0.0596386788 |
transcription factor |
TF Family: NAC |
hypothetical protein POPTR_0015s03700g [Populus trichocarpa] |
| 15 |
Hb_002944_020 |
0.0627459966 |
transcription factor |
TF Family: bZIP |
hypothetical protein POPTR_0006s05770g, partial [Populus trichocarpa] |
| 16 |
Hb_000500_190 |
0.0654571113 |
- |
- |
hypothetical protein JCGZ_21655 [Jatropha curcas] |
| 17 |
Hb_005464_040 |
0.0661417149 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 18 |
Hb_000589_280 |
0.0662957169 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase 5 [Jatropha curcas] |
| 19 |
Hb_004981_020 |
0.0676164992 |
- |
- |
PREDICTED: uncharacterized protein LOC103706119 [Phoenix dactylifera] |
| 20 |
Hb_143766_040 |
0.067959245 |
- |
- |
PREDICTED: probable caffeoyl-CoA O-methyltransferase At4g26220 [Jatropha curcas] |