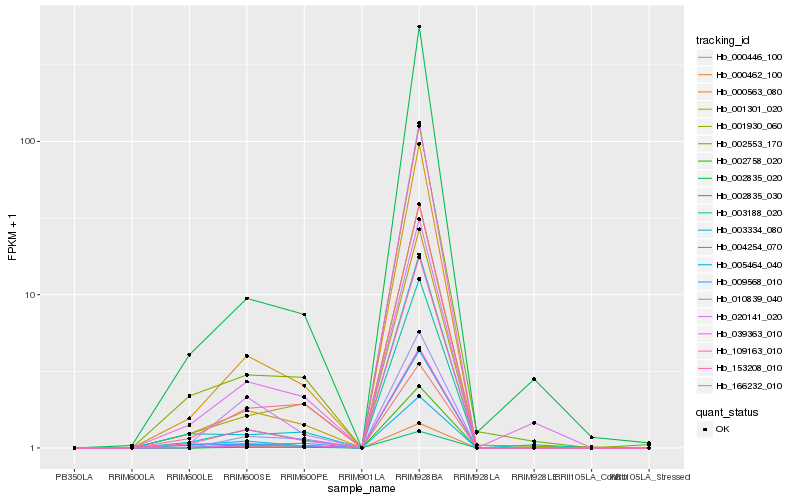
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_166232_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_22831 [Jatropha curcas] |
| 2 |
Hb_000563_080 |
0.0368945331 |
- |
- |
- |
| 3 |
Hb_001930_060 |
0.0381281829 |
- |
- |
PREDICTED: cytochrome P450 71D9-like [Fragaria vesca subsp. vesca] |
| 4 |
Hb_001301_020 |
0.04159361 |
- |
- |
hypothetical protein JCGZ_06575 [Jatropha curcas] |
| 5 |
Hb_002758_020 |
0.0442922276 |
- |
- |
PREDICTED: putative auxin efflux carrier component 5 [Jatropha curcas] |
| 6 |
Hb_003188_020 |
0.0504159005 |
- |
- |
chitinase, putative [Ricinus communis] |
| 7 |
Hb_002835_030 |
0.0528427518 |
- |
- |
PREDICTED: lysM domain receptor-like kinase 4 [Vitis vinifera] |
| 8 |
Hb_005464_040 |
0.0549221412 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 9 |
Hb_002553_170 |
0.0594123294 |
- |
- |
PREDICTED: early nodulin-75-like [Jatropha curcas] |
| 10 |
Hb_153208_010 |
0.0607756358 |
- |
- |
Cationic peroxidase 2 precursor, putative [Ricinus communis] |
| 11 |
Hb_010839_040 |
0.0623791911 |
- |
- |
hypothetical protein JCGZ_20868 [Jatropha curcas] |
| 12 |
Hb_009568_010 |
0.0625192117 |
- |
- |
hypothetical protein POPTR_0001s46700g [Populus trichocarpa] |
| 13 |
Hb_109163_010 |
0.0637104492 |
- |
- |
polyphenol oxidase [Hevea brasiliensis] |
| 14 |
Hb_002835_020 |
0.0645195045 |
- |
- |
PREDICTED: cytochrome P450 71A1-like [Jatropha curcas] |
| 15 |
Hb_000446_100 |
0.0651053098 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 16 |
Hb_000462_100 |
0.0664988363 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 17 |
Hb_003334_080 |
0.0690010509 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 18 |
Hb_039363_010 |
0.0714181828 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 19 |
Hb_020141_020 |
0.0723492397 |
- |
- |
- |
| 20 |
Hb_004254_070 |
0.0728674567 |
transcription factor |
TF Family: NAC |
hypothetical protein POPTR_0015s03700g [Populus trichocarpa] |