| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
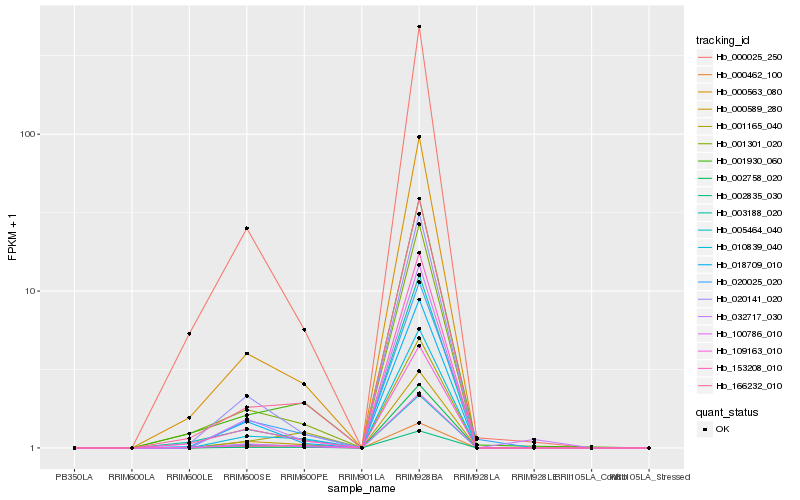
Hb_002835_030 |
0.0 |
- |
- |
PREDICTED: lysM domain receptor-like kinase 4 [Vitis vinifera] |
| 2 |
Hb_010839_040 |
0.0160208668 |
- |
- |
hypothetical protein JCGZ_20868 [Jatropha curcas] |
| 3 |
Hb_000462_100 |
0.020982773 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 4 |
Hb_002758_020 |
0.0252027607 |
- |
- |
PREDICTED: putative auxin efflux carrier component 5 [Jatropha curcas] |
| 5 |
Hb_000589_280 |
0.0366838423 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase 5 [Jatropha curcas] |
| 6 |
Hb_166232_010 |
0.0528427518 |
- |
- |
hypothetical protein JCGZ_22831 [Jatropha curcas] |
| 7 |
Hb_100786_010 |
0.0559270347 |
- |
- |
hypothetical protein B456_008G202600 [Gossypium raimondii] |
| 8 |
Hb_000563_080 |
0.0649100156 |
- |
- |
- |
| 9 |
Hb_001165_040 |
0.0665514327 |
- |
- |
hypothetical protein POPTR_0009s10120g [Populus trichocarpa] |
| 10 |
Hb_005464_040 |
0.0685933549 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 11 |
Hb_001301_020 |
0.0762126391 |
- |
- |
hypothetical protein JCGZ_06575 [Jatropha curcas] |
| 12 |
Hb_001930_060 |
0.076933668 |
- |
- |
PREDICTED: cytochrome P450 71D9-like [Fragaria vesca subsp. vesca] |
| 13 |
Hb_020141_020 |
0.0792267737 |
- |
- |
- |
| 14 |
Hb_003188_020 |
0.0839066981 |
- |
- |
chitinase, putative [Ricinus communis] |
| 15 |
Hb_020025_020 |
0.0873998201 |
- |
- |
SENESCENCE-RELATED GENE 1 family protein [Populus trichocarpa] |
| 16 |
Hb_153208_010 |
0.090920724 |
- |
- |
Cationic peroxidase 2 precursor, putative [Ricinus communis] |
| 17 |
Hb_018709_010 |
0.0910888751 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 18 |
Hb_032717_030 |
0.0974520055 |
- |
- |
PREDICTED: CASP-like protein 1B2 [Jatropha curcas] |
| 19 |
Hb_000025_250 |
0.0976330145 |
- |
- |
PREDICTED: flavonoid 3',5'-hydroxylase 2-like [Jatropha curcas] |
| 20 |
Hb_109163_010 |
0.0977268645 |
- |
- |
polyphenol oxidase [Hevea brasiliensis] |