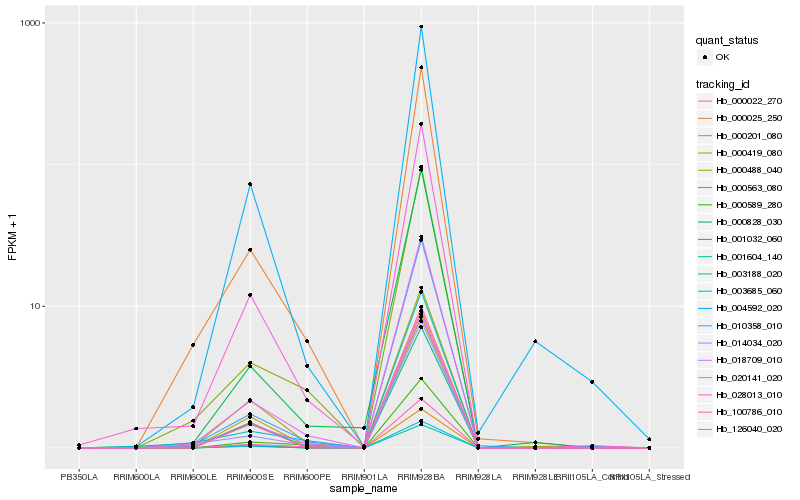
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_028013_010 |
0.0 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 2 |
Hb_018709_010 |
0.0368978316 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 3 |
Hb_020141_020 |
0.0501361629 |
- |
- |
- |
| 4 |
Hb_001032_060 |
0.0579986577 |
- |
- |
PREDICTED: ankyrin-2-like [Citrus sinensis] |
| 5 |
Hb_000025_250 |
0.0580336883 |
- |
- |
PREDICTED: flavonoid 3',5'-hydroxylase 2-like [Jatropha curcas] |
| 6 |
Hb_100786_010 |
0.0606682541 |
- |
- |
hypothetical protein B456_008G202600 [Gossypium raimondii] |
| 7 |
Hb_000419_080 |
0.068242047 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-4 [Jatropha curcas] |
| 8 |
Hb_000201_080 |
0.071376087 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 9 |
Hb_001604_140 |
0.0713850322 |
- |
- |
PREDICTED: ABC transporter G family member 10-like [Populus euphratica] |
| 10 |
Hb_003685_060 |
0.0715393043 |
- |
- |
PREDICTED: laccase-15 [Jatropha curcas] |
| 11 |
Hb_126040_020 |
0.0717044277 |
- |
- |
- |
| 12 |
Hb_000022_270 |
0.0717733719 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000488_040 |
0.0733852145 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 14 |
Hb_004592_020 |
0.0736962751 |
- |
- |
PREDICTED: non-specific lipid-transfer protein 1-like [Jatropha curcas] |
| 15 |
Hb_014034_020 |
0.0750615302 |
- |
- |
hypothetical protein POPTR_0075s00200g, partial [Populus trichocarpa] |
| 16 |
Hb_000828_030 |
0.0761259847 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 17 |
Hb_000563_080 |
0.0774890183 |
- |
- |
- |
| 18 |
Hb_010358_010 |
0.0781961853 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 19 |
Hb_000589_280 |
0.0789957413 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase 5 [Jatropha curcas] |
| 20 |
Hb_003188_020 |
0.0806645385 |
- |
- |
chitinase, putative [Ricinus communis] |