| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
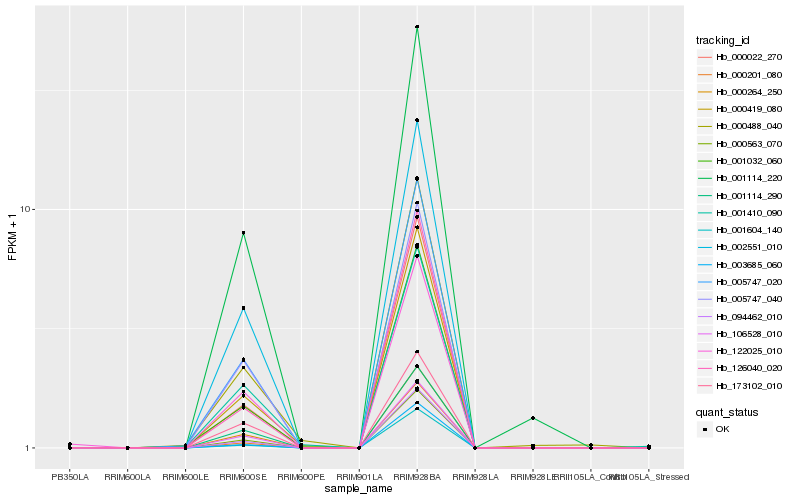
Hb_000563_070 |
0.0 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase LOG2 [Sesamum indicum] |
| 2 |
Hb_000264_250 |
0.0105629247 |
- |
- |
thioredoxin family protein [Arabidopsis thaliana] |
| 3 |
Hb_002551_010 |
0.0214328514 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 4 |
Hb_005747_020 |
0.0269499627 |
- |
- |
bacterial-induced peroxidase [Gossypium schwendimanii] |
| 5 |
Hb_094462_010 |
0.0272670922 |
- |
- |
hypothetical protein CISIN_1g033698mg [Citrus sinensis] |
| 6 |
Hb_106528_010 |
0.0311061176 |
- |
- |
Cationic peroxidase 2 precursor, putative [Ricinus communis] |
| 7 |
Hb_005747_040 |
0.0326166306 |
- |
- |
PREDICTED: peroxidase N1-like, partial [Nicotiana sylvestris] |
| 8 |
Hb_000419_080 |
0.044469321 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-4 [Jatropha curcas] |
| 9 |
Hb_001114_290 |
0.0515214166 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001114_220 |
0.0535789502 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 11 |
Hb_001032_060 |
0.0570680577 |
- |
- |
PREDICTED: ankyrin-2-like [Citrus sinensis] |
| 12 |
Hb_000022_270 |
0.0605243958 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_003685_060 |
0.0630560361 |
- |
- |
PREDICTED: laccase-15 [Jatropha curcas] |
| 14 |
Hb_122025_010 |
0.0631666279 |
- |
- |
hypothetical protein POPTR_0003s052201g, partial [Populus trichocarpa] |
| 15 |
Hb_001604_140 |
0.0658654916 |
- |
- |
PREDICTED: ABC transporter G family member 10-like [Populus euphratica] |
| 16 |
Hb_173102_010 |
0.069840446 |
- |
- |
Ent-kaurene synthase B, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_000201_080 |
0.0705146055 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 18 |
Hb_001410_090 |
0.0721591645 |
- |
- |
transferase, putative [Ricinus communis] |
| 19 |
Hb_126040_020 |
0.0754538011 |
- |
- |
- |
| 20 |
Hb_000488_040 |
0.076377062 |
- |
- |
unnamed protein product [Vitis vinifera] |