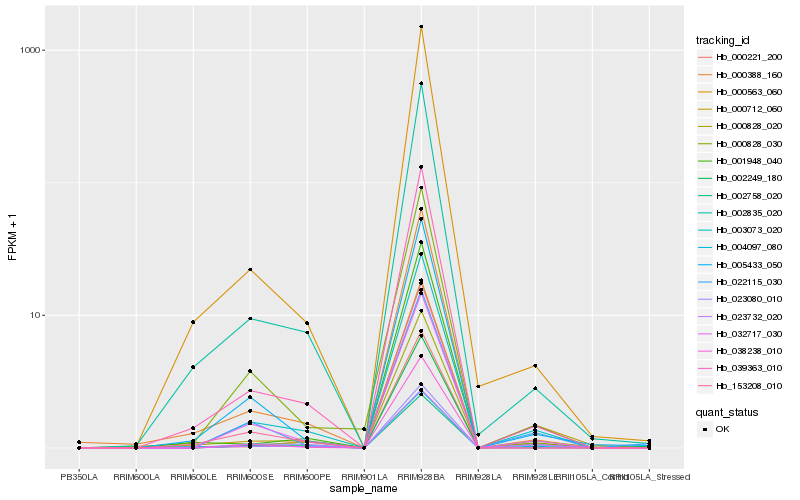
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003073_020 |
0.0 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 2 |
Hb_032717_030 |
0.0486177316 |
- |
- |
PREDICTED: CASP-like protein 1B2 [Jatropha curcas] |
| 3 |
Hb_023732_020 |
0.0574263077 |
- |
- |
PREDICTED: fatty acyl-CoA reductase 3-like [Jatropha curcas] |
| 4 |
Hb_039363_010 |
0.0646188814 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 5 |
Hb_000828_020 |
0.0693123636 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 6 |
Hb_002835_020 |
0.0741618997 |
- |
- |
PREDICTED: cytochrome P450 71A1-like [Jatropha curcas] |
| 7 |
Hb_005433_050 |
0.0777592104 |
- |
- |
PREDICTED: uncharacterized protein At1g66480-like [Populus euphratica] |
| 8 |
Hb_023080_010 |
0.0784195057 |
- |
- |
PREDICTED: probable serine incorporator [Jatropha curcas] |
| 9 |
Hb_002249_180 |
0.0785485838 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 10 |
Hb_000388_160 |
0.0823071157 |
- |
- |
xyloglucan endo-1 family protein [Populus trichocarpa] |
| 11 |
Hb_000221_200 |
0.0838344529 |
- |
- |
hypothetical protein JCGZ_15819 [Jatropha curcas] |
| 12 |
Hb_000712_060 |
0.0855474681 |
- |
- |
PREDICTED: uncharacterized protein LOC105631517 [Jatropha curcas] |
| 13 |
Hb_022115_030 |
0.0856630593 |
- |
- |
PREDICTED: umecyanin-like [Jatropha curcas] |
| 14 |
Hb_000563_060 |
0.0870298121 |
- |
- |
PREDICTED: uncharacterized protein LOC100306381 isoform X1 [Glycine max] |
| 15 |
Hb_002758_020 |
0.0874983111 |
- |
- |
PREDICTED: putative auxin efflux carrier component 5 [Jatropha curcas] |
| 16 |
Hb_001948_040 |
0.0921732153 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranyl geranyl pyrophosphate synthase, putative [Ricinus communis] |
| 17 |
Hb_153208_010 |
0.092298595 |
- |
- |
Cationic peroxidase 2 precursor, putative [Ricinus communis] |
| 18 |
Hb_038238_010 |
0.0924033438 |
- |
- |
hypothetical protein POPTR_0001s30980g [Populus trichocarpa] |
| 19 |
Hb_000828_030 |
0.0926214926 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 20 |
Hb_004097_080 |
0.0937820215 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 isoform X1 [Nicotiana tomentosiformis] |