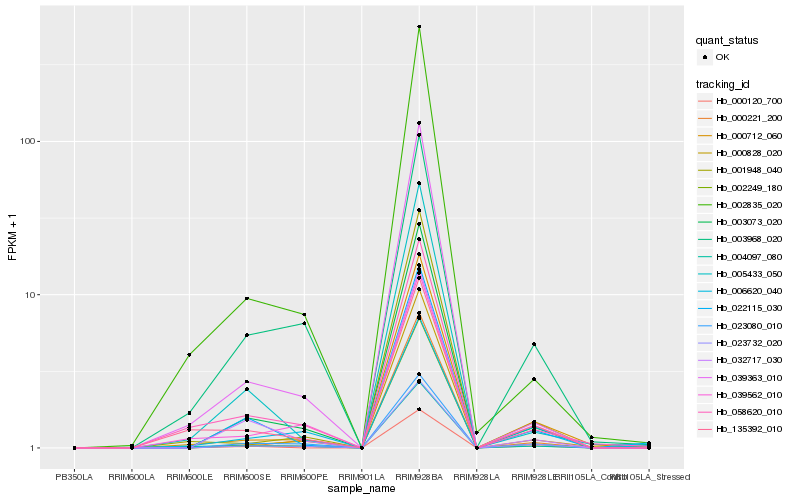
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_023732_020 |
0.0 |
- |
- |
PREDICTED: fatty acyl-CoA reductase 3-like [Jatropha curcas] |
| 2 |
Hb_003073_020 |
0.0574263077 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 3 |
Hb_023080_010 |
0.0635296819 |
- |
- |
PREDICTED: probable serine incorporator [Jatropha curcas] |
| 4 |
Hb_000712_060 |
0.071625554 |
- |
- |
PREDICTED: uncharacterized protein LOC105631517 [Jatropha curcas] |
| 5 |
Hb_000828_020 |
0.0845765155 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 6 |
Hb_032717_030 |
0.0889837701 |
- |
- |
PREDICTED: CASP-like protein 1B2 [Jatropha curcas] |
| 7 |
Hb_039562_010 |
0.09010305 |
- |
- |
PREDICTED: protein SRG1-like [Jatropha curcas] |
| 8 |
Hb_000120_700 |
0.0937326046 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 9 |
Hb_000221_200 |
0.0946947004 |
- |
- |
hypothetical protein JCGZ_15819 [Jatropha curcas] |
| 10 |
Hb_022115_030 |
0.0954195937 |
- |
- |
PREDICTED: umecyanin-like [Jatropha curcas] |
| 11 |
Hb_002249_180 |
0.1017423494 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 12 |
Hb_004097_080 |
0.1029896071 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 isoform X1 [Nicotiana tomentosiformis] |
| 13 |
Hb_001948_040 |
0.1033925627 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranyl geranyl pyrophosphate synthase, putative [Ricinus communis] |
| 14 |
Hb_039363_010 |
0.1081254574 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 15 |
Hb_135392_010 |
0.1120814382 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase-like [Jatropha curcas] |
| 16 |
Hb_006620_040 |
0.1133721283 |
- |
- |
PREDICTED: probable 6-phosphogluconolactonase 1 [Jatropha curcas] |
| 17 |
Hb_003968_020 |
0.1137896284 |
- |
- |
PREDICTED: limonoid UDP-glucosyltransferase-like [Glycine max] |
| 18 |
Hb_002835_020 |
0.1145070772 |
- |
- |
PREDICTED: cytochrome P450 71A1-like [Jatropha curcas] |
| 19 |
Hb_058620_010 |
0.1148314358 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 20 |
Hb_005433_050 |
0.1174312463 |
- |
- |
PREDICTED: uncharacterized protein At1g66480-like [Populus euphratica] |