| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
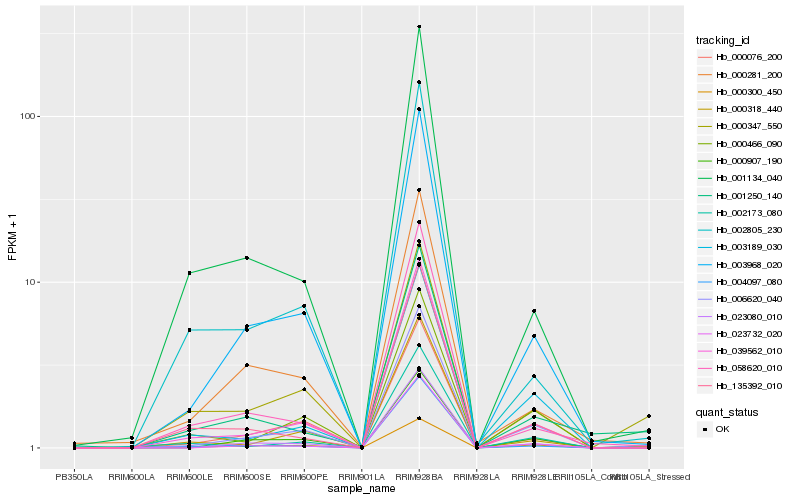
Hb_006620_040 |
0.0 |
- |
- |
PREDICTED: probable 6-phosphogluconolactonase 1 [Jatropha curcas] |
| 2 |
Hb_003968_020 |
0.0766630459 |
- |
- |
PREDICTED: limonoid UDP-glucosyltransferase-like [Glycine max] |
| 3 |
Hb_039562_010 |
0.0879798827 |
- |
- |
PREDICTED: protein SRG1-like [Jatropha curcas] |
| 4 |
Hb_000466_090 |
0.0967615292 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RGLG2-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_023080_010 |
0.0985918256 |
- |
- |
PREDICTED: probable serine incorporator [Jatropha curcas] |
| 6 |
Hb_003189_030 |
0.0998811875 |
- |
- |
hypothetical protein Csa_4G304750 [Cucumis sativus] |
| 7 |
Hb_000907_190 |
0.1013307197 |
- |
- |
PREDICTED: uncharacterized protein At4g15970-like [Jatropha curcas] |
| 8 |
Hb_023732_020 |
0.1133721283 |
- |
- |
PREDICTED: fatty acyl-CoA reductase 3-like [Jatropha curcas] |
| 9 |
Hb_000076_200 |
0.1137370761 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000318_440 |
0.1198439474 |
- |
- |
PREDICTED: allene oxide synthase 2-like [Jatropha curcas] |
| 11 |
Hb_002173_080 |
0.1213744806 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 12 |
Hb_000300_450 |
0.1280752 |
- |
- |
PREDICTED: caffeic acid 3-O-methyltransferase-like isoform X1 [Citrus sinensis] |
| 13 |
Hb_135392_010 |
0.1300838506 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase-like [Jatropha curcas] |
| 14 |
Hb_000281_200 |
0.1336560576 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 15 |
Hb_058620_010 |
0.1345230772 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 16 |
Hb_002805_230 |
0.1382779229 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001134_040 |
0.1385285089 |
- |
- |
PREDICTED: EG45-like domain containing protein [Jatropha curcas] |
| 18 |
Hb_001250_140 |
0.1387109867 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 2 [Jatropha curcas] |
| 19 |
Hb_000347_550 |
0.139153807 |
transcription factor |
TF Family: MYB |
Duplicated homeodomain-like superfamily protein, putative [Theobroma cacao] |
| 20 |
Hb_004097_080 |
0.140360811 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 isoform X1 [Nicotiana tomentosiformis] |