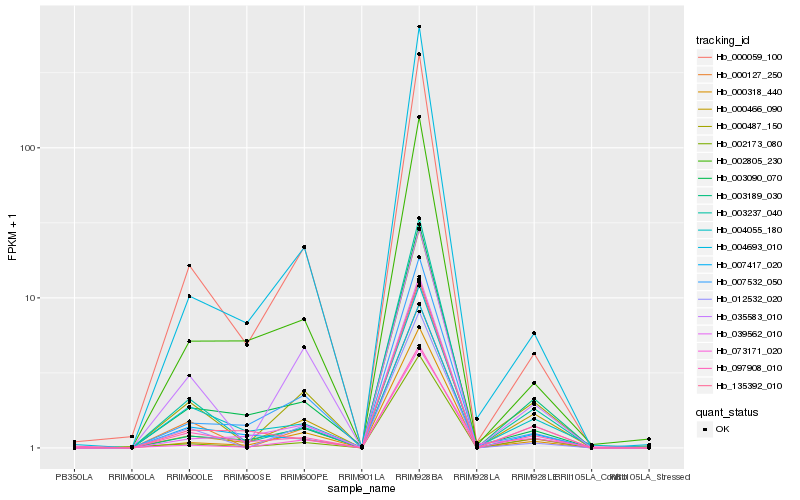
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000487_150 |
0.0 |
- |
- |
PREDICTED: feruloyl CoA ortho-hydroxylase 2-like [Jatropha curcas] |
| 2 |
Hb_000318_440 |
0.082622563 |
- |
- |
PREDICTED: allene oxide synthase 2-like [Jatropha curcas] |
| 3 |
Hb_012532_020 |
0.0886582904 |
- |
- |
Isoflavone reductase, putative [Ricinus communis] |
| 4 |
Hb_000127_250 |
0.0914614595 |
- |
- |
Alliin lyase precursor, putative [Ricinus communis] |
| 5 |
Hb_002173_080 |
0.0954475522 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 6 |
Hb_000059_100 |
0.0971157313 |
- |
- |
Basic blue protein, putative [Ricinus communis] |
| 7 |
Hb_039562_010 |
0.1029658889 |
- |
- |
PREDICTED: protein SRG1-like [Jatropha curcas] |
| 8 |
Hb_003237_040 |
0.1041566888 |
- |
- |
Ent-kaurene synthase B, chloroplast precursor, putative [Ricinus communis] |
| 9 |
Hb_007532_050 |
0.1081132325 |
- |
- |
9-cis-epoxycarotenoid dioxygenase, putative [Ricinus communis] |
| 10 |
Hb_000466_090 |
0.110638021 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RGLG2-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_004693_010 |
0.1147113715 |
- |
- |
hypothetical protein JCGZ_02321 [Jatropha curcas] |
| 12 |
Hb_003090_070 |
0.1147602587 |
- |
- |
PREDICTED: laccase-7 [Jatropha curcas] |
| 13 |
Hb_073171_020 |
0.1156446731 |
- |
- |
PREDICTED: ABC transporter G family member 29-like [Jatropha curcas] |
| 14 |
Hb_035583_010 |
0.1218489056 |
- |
- |
PREDICTED: premnaspirodiene oxygenase-like [Jatropha curcas] |
| 15 |
Hb_004055_180 |
0.1228945655 |
- |
- |
cell wall invertase [Manihot esculenta] |
| 16 |
Hb_002805_230 |
0.1233326005 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12 isoform X1 [Jatropha curcas] |
| 17 |
Hb_007417_020 |
0.1237229541 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 18 |
Hb_097908_010 |
0.124387786 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g03230 isoform X2 [Jatropha curcas] |
| 19 |
Hb_003189_030 |
0.1268114708 |
- |
- |
hypothetical protein Csa_4G304750 [Cucumis sativus] |
| 20 |
Hb_135392_010 |
0.1328941263 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase-like [Jatropha curcas] |