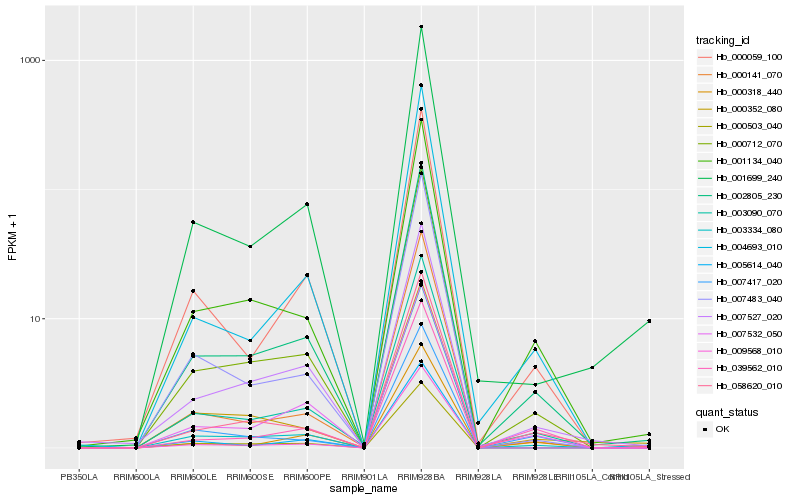
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002805_230 |
0.0 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12 isoform X1 [Jatropha curcas] |
| 2 |
Hb_003090_070 |
0.0386110184 |
- |
- |
PREDICTED: laccase-7 [Jatropha curcas] |
| 3 |
Hb_000712_070 |
0.0431113643 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001134_040 |
0.0476715725 |
- |
- |
PREDICTED: EG45-like domain containing protein [Jatropha curcas] |
| 5 |
Hb_007532_050 |
0.0648229378 |
- |
- |
9-cis-epoxycarotenoid dioxygenase, putative [Ricinus communis] |
| 6 |
Hb_058620_010 |
0.0653303965 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 7 |
Hb_004693_010 |
0.0685926906 |
- |
- |
hypothetical protein JCGZ_02321 [Jatropha curcas] |
| 8 |
Hb_000059_100 |
0.0688472025 |
- |
- |
Basic blue protein, putative [Ricinus communis] |
| 9 |
Hb_001699_240 |
0.070275011 |
- |
- |
PREDICTED: naringenin,2-oxoglutarate 3-dioxygenase [Jatropha curcas] |
| 10 |
Hb_007527_020 |
0.0710963616 |
- |
- |
PREDICTED: feruloyl CoA ortho-hydroxylase 1 [Jatropha curcas] |
| 11 |
Hb_000318_440 |
0.0787111921 |
- |
- |
PREDICTED: allene oxide synthase 2-like [Jatropha curcas] |
| 12 |
Hb_000503_040 |
0.0800654336 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase-like [Jatropha curcas] |
| 13 |
Hb_000141_070 |
0.0811067137 |
- |
- |
l-lactate dehydrogenase, putative [Ricinus communis] |
| 14 |
Hb_007483_040 |
0.0812103112 |
- |
- |
PREDICTED: protein SRG1-like [Jatropha curcas] |
| 15 |
Hb_039562_010 |
0.0842409024 |
- |
- |
PREDICTED: protein SRG1-like [Jatropha curcas] |
| 16 |
Hb_000352_080 |
0.0877182401 |
- |
- |
PREDICTED: lipid transfer-like protein VAS [Jatropha curcas] |
| 17 |
Hb_007417_020 |
0.0897425998 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 18 |
Hb_003334_080 |
0.0906793249 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 19 |
Hb_005614_040 |
0.0916430911 |
- |
- |
unnamed protein product [Coffea canephora] |
| 20 |
Hb_009568_010 |
0.092175283 |
- |
- |
hypothetical protein POPTR_0001s46700g [Populus trichocarpa] |