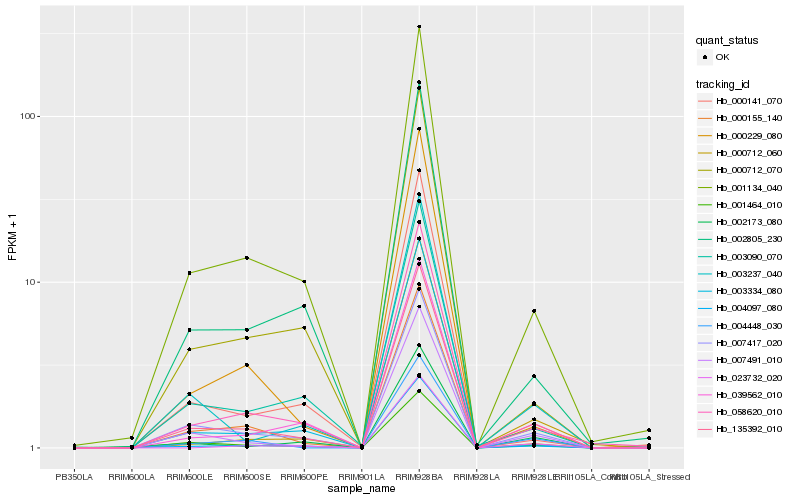
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_135392_010 |
0.0 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase-like [Jatropha curcas] |
| 2 |
Hb_007417_020 |
0.0483007251 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 3 |
Hb_007491_010 |
0.059241291 |
- |
- |
Spotted leaf protein, putative [Ricinus communis] |
| 4 |
Hb_001134_040 |
0.0726009372 |
- |
- |
PREDICTED: EG45-like domain containing protein [Jatropha curcas] |
| 5 |
Hb_058620_010 |
0.0756460203 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 6 |
Hb_001464_010 |
0.0782295919 |
- |
- |
PREDICTED: putative receptor-like protein kinase At4g00960 isoform X1 [Populus euphratica] |
| 7 |
Hb_004097_080 |
0.0788438204 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 isoform X1 [Nicotiana tomentosiformis] |
| 8 |
Hb_039562_010 |
0.078990309 |
- |
- |
PREDICTED: protein SRG1-like [Jatropha curcas] |
| 9 |
Hb_003237_040 |
0.0889754931 |
- |
- |
Ent-kaurene synthase B, chloroplast precursor, putative [Ricinus communis] |
| 10 |
Hb_004448_030 |
0.0912966635 |
- |
- |
PREDICTED: RING-H2 finger protein ATL40-like [Jatropha curcas] |
| 11 |
Hb_003090_070 |
0.0957987721 |
- |
- |
PREDICTED: laccase-7 [Jatropha curcas] |
| 12 |
Hb_002805_230 |
0.0984233456 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000712_070 |
0.0994694808 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000155_140 |
0.100554315 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 15 |
Hb_002173_080 |
0.1040444689 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 16 |
Hb_000229_080 |
0.1061714706 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 17 |
Hb_000712_060 |
0.1111411361 |
- |
- |
PREDICTED: uncharacterized protein LOC105631517 [Jatropha curcas] |
| 18 |
Hb_023732_020 |
0.1120814382 |
- |
- |
PREDICTED: fatty acyl-CoA reductase 3-like [Jatropha curcas] |
| 19 |
Hb_003334_080 |
0.1137779135 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 20 |
Hb_000141_070 |
0.1150894941 |
- |
- |
l-lactate dehydrogenase, putative [Ricinus communis] |