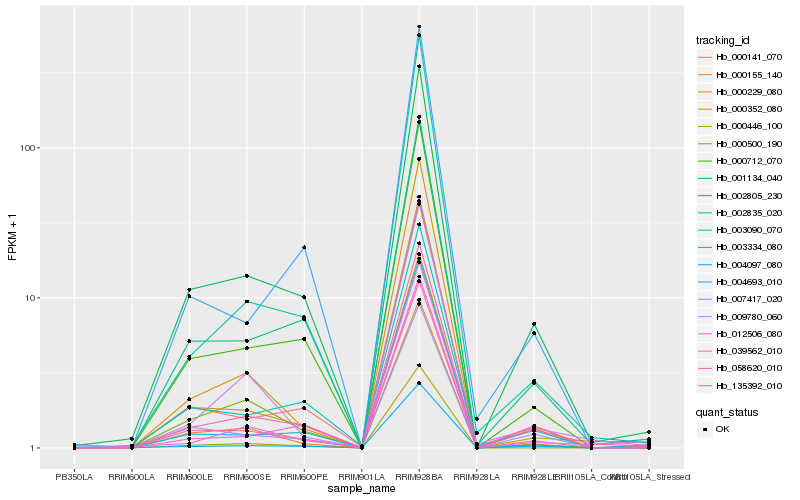
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_058620_010 |
0.0 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 2 |
Hb_001134_040 |
0.0565132443 |
- |
- |
PREDICTED: EG45-like domain containing protein [Jatropha curcas] |
| 3 |
Hb_000712_070 |
0.0644542354 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002805_230 |
0.0653303965 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12 isoform X1 [Jatropha curcas] |
| 5 |
Hb_004097_080 |
0.0710895696 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 isoform X1 [Nicotiana tomentosiformis] |
| 6 |
Hb_135392_010 |
0.0756460203 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase-like [Jatropha curcas] |
| 7 |
Hb_003090_070 |
0.0782478945 |
- |
- |
PREDICTED: laccase-7 [Jatropha curcas] |
| 8 |
Hb_000155_140 |
0.0812857655 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 9 |
Hb_003334_080 |
0.0830158569 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 10 |
Hb_000141_070 |
0.0832355987 |
- |
- |
l-lactate dehydrogenase, putative [Ricinus communis] |
| 11 |
Hb_000352_080 |
0.0838334334 |
- |
- |
PREDICTED: lipid transfer-like protein VAS [Jatropha curcas] |
| 12 |
Hb_009780_060 |
0.0838806331 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranyl geranyl pyrophosphate synthase, putative [Ricinus communis] |
| 13 |
Hb_039562_010 |
0.0841263542 |
- |
- |
PREDICTED: protein SRG1-like [Jatropha curcas] |
| 14 |
Hb_000229_080 |
0.0851392976 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 15 |
Hb_002835_020 |
0.0870731379 |
- |
- |
PREDICTED: cytochrome P450 71A1-like [Jatropha curcas] |
| 16 |
Hb_004693_010 |
0.0882555425 |
- |
- |
hypothetical protein JCGZ_02321 [Jatropha curcas] |
| 17 |
Hb_000500_190 |
0.0907107013 |
- |
- |
hypothetical protein JCGZ_21655 [Jatropha curcas] |
| 18 |
Hb_007417_020 |
0.0910770535 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 19 |
Hb_012506_080 |
0.0941600499 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 20 |
Hb_000446_100 |
0.0942821857 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |