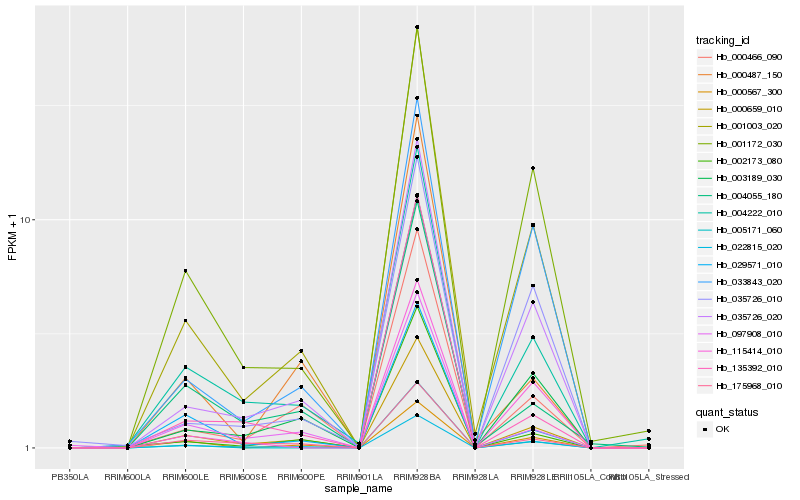
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001003_020 |
0.0 |
- |
- |
PREDICTED: metalloendoproteinase 1-like [Populus euphratica] |
| 2 |
Hb_035726_020 |
0.0603440748 |
- |
- |
Insulinase (Peptidase family M16) family protein [Theobroma cacao] |
| 3 |
Hb_003189_030 |
0.0678798142 |
- |
- |
hypothetical protein Csa_4G304750 [Cucumis sativus] |
| 4 |
Hb_000659_010 |
0.0744297154 |
- |
- |
PREDICTED: plasma membrane ATPase 2-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_022815_020 |
0.078995508 |
- |
- |
PREDICTED: uncharacterized protein LOC105650107 [Jatropha curcas] |
| 6 |
Hb_004222_010 |
0.1017999737 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Jatropha curcas] |
| 7 |
Hb_033843_020 |
0.1096149053 |
- |
- |
lysosomal alpha-mannosidase, putative [Ricinus communis] |
| 8 |
Hb_000466_090 |
0.1113355334 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RGLG2-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_001172_030 |
0.1160156664 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 7.2-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_002173_080 |
0.1230623266 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 11 |
Hb_035726_010 |
0.1241525466 |
- |
- |
Insulin-degrading enzyme, putative [Ricinus communis] |
| 12 |
Hb_175968_010 |
0.1329477164 |
- |
- |
hypothetical protein JCGZ_21281 [Jatropha curcas] |
| 13 |
Hb_115414_010 |
0.13299959 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Jatropha curcas] |
| 14 |
Hb_000487_150 |
0.138603351 |
- |
- |
PREDICTED: feruloyl CoA ortho-hydroxylase 2-like [Jatropha curcas] |
| 15 |
Hb_005171_060 |
0.1397559979 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 16 |
Hb_097908_010 |
0.1409872392 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g03230 isoform X2 [Jatropha curcas] |
| 17 |
Hb_004055_180 |
0.1417458838 |
- |
- |
cell wall invertase [Manihot esculenta] |
| 18 |
Hb_000567_300 |
0.1438213528 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 19 |
Hb_029571_010 |
0.1449087627 |
- |
- |
PREDICTED: polygalacturonase QRT3 [Jatropha curcas] |
| 20 |
Hb_135392_010 |
0.1476132103 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase-like [Jatropha curcas] |