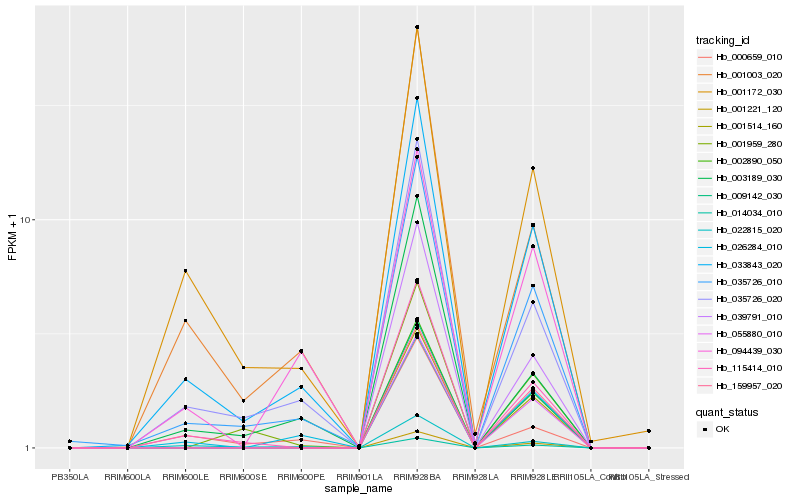
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_035726_010 |
0.0 |
- |
- |
Insulin-degrading enzyme, putative [Ricinus communis] |
| 2 |
Hb_033843_020 |
0.0656930195 |
- |
- |
lysosomal alpha-mannosidase, putative [Ricinus communis] |
| 3 |
Hb_035726_020 |
0.0869237604 |
- |
- |
Insulinase (Peptidase family M16) family protein [Theobroma cacao] |
| 4 |
Hb_115414_010 |
0.1069084646 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Jatropha curcas] |
| 5 |
Hb_001172_030 |
0.1188882723 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 7.2-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_022815_020 |
0.1192023441 |
- |
- |
PREDICTED: uncharacterized protein LOC105650107 [Jatropha curcas] |
| 7 |
Hb_001003_020 |
0.1241525466 |
- |
- |
PREDICTED: metalloendoproteinase 1-like [Populus euphratica] |
| 8 |
Hb_001959_280 |
0.1308190842 |
- |
- |
PREDICTED: uncharacterized protein LOC105638432 [Jatropha curcas] |
| 9 |
Hb_003189_030 |
0.1359776862 |
- |
- |
hypothetical protein Csa_4G304750 [Cucumis sativus] |
| 10 |
Hb_026284_010 |
0.1423669905 |
- |
- |
- |
| 11 |
Hb_001221_120 |
0.1499629779 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 12 |
Hb_002890_050 |
0.1504607947 |
- |
- |
- |
| 13 |
Hb_014034_010 |
0.150667478 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 14 |
Hb_094439_030 |
0.1508680873 |
- |
- |
PREDICTED: probable S-adenosylmethionine-dependent methyltransferase At5g38780 [Eucalyptus grandis] |
| 15 |
Hb_001514_160 |
0.1517493809 |
- |
- |
hypothetical protein JCGZ_16070 [Jatropha curcas] |
| 16 |
Hb_009142_030 |
0.1522360631 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 17 |
Hb_039791_010 |
0.1529901262 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_055880_010 |
0.1530330254 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 19 |
Hb_000659_010 |
0.1602169346 |
- |
- |
PREDICTED: plasma membrane ATPase 2-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_159957_020 |
0.160981219 |
- |
- |
- |