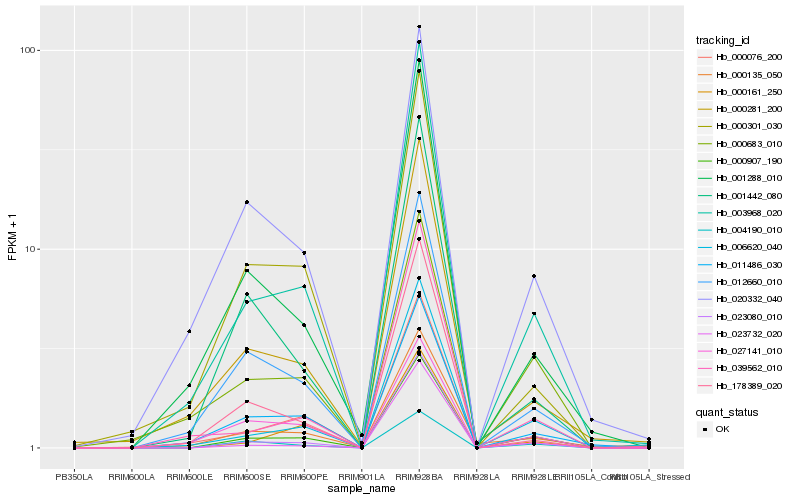
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000907_190 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At4g15970-like [Jatropha curcas] |
| 2 |
Hb_003968_020 |
0.0929796077 |
- |
- |
PREDICTED: limonoid UDP-glucosyltransferase-like [Glycine max] |
| 3 |
Hb_023080_010 |
0.1004188621 |
- |
- |
PREDICTED: probable serine incorporator [Jatropha curcas] |
| 4 |
Hb_006620_040 |
0.1013307197 |
- |
- |
PREDICTED: probable 6-phosphogluconolactonase 1 [Jatropha curcas] |
| 5 |
Hb_004190_010 |
0.1093920445 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_000076_200 |
0.1185151861 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_012660_010 |
0.119865901 |
- |
- |
hypothetical protein JCGZ_25971 [Jatropha curcas] |
| 8 |
Hb_011486_030 |
0.1211611828 |
- |
- |
branched-chain amino acid aminotransferase, putative [Ricinus communis] |
| 9 |
Hb_000161_250 |
0.1322366586 |
transcription factor |
TF Family: Tify |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001288_010 |
0.1337201872 |
- |
- |
- |
| 11 |
Hb_020332_040 |
0.1360590494 |
- |
- |
PREDICTED: alcohol dehydrogenase 1-like [Jatropha curcas] |
| 12 |
Hb_000683_010 |
0.1392932085 |
- |
- |
Interleukin-1 receptor-associated kinase, putative [Ricinus communis] |
| 13 |
Hb_000281_200 |
0.1425135087 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 14 |
Hb_001442_080 |
0.143033642 |
transcription factor |
TF Family: MYB-related |
transcription factor MYB251 [Fagus crenata] |
| 15 |
Hb_023732_020 |
0.1449665774 |
- |
- |
PREDICTED: fatty acyl-CoA reductase 3-like [Jatropha curcas] |
| 16 |
Hb_039562_010 |
0.1452212522 |
- |
- |
PREDICTED: protein SRG1-like [Jatropha curcas] |
| 17 |
Hb_000301_030 |
0.1468031995 |
- |
- |
PREDICTED: pectate lyase-like [Jatropha curcas] |
| 18 |
Hb_027141_010 |
0.1476519212 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 19 |
Hb_000135_050 |
0.1486577883 |
- |
- |
PREDICTED: putative leucine-rich repeat receptor-like protein kinase At2g19210-like isoform X2 [Citrus sinensis] |
| 20 |
Hb_178389_020 |
0.1521443975 |
- |
- |
PREDICTED: cytochrome P450 86B1-like [Jatropha curcas] |