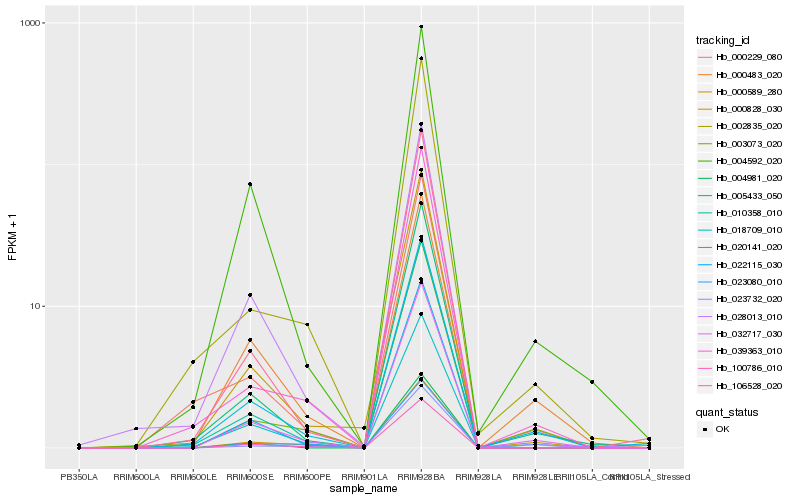
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_032717_030 |
0.0 |
- |
- |
PREDICTED: CASP-like protein 1B2 [Jatropha curcas] |
| 2 |
Hb_003073_020 |
0.0486177316 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 3 |
Hb_005433_050 |
0.0657155807 |
- |
- |
PREDICTED: uncharacterized protein At1g66480-like [Populus euphratica] |
| 4 |
Hb_022115_030 |
0.0692946489 |
- |
- |
PREDICTED: umecyanin-like [Jatropha curcas] |
| 5 |
Hb_020141_020 |
0.074871908 |
- |
- |
- |
| 6 |
Hb_100786_010 |
0.075881334 |
- |
- |
hypothetical protein B456_008G202600 [Gossypium raimondii] |
| 7 |
Hb_000828_030 |
0.076934621 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 8 |
Hb_000483_020 |
0.0804789586 |
- |
- |
PREDICTED: 7-deoxyloganetic acid glucosyltransferase-like isoform X2 [Jatropha curcas] |
| 9 |
Hb_039363_010 |
0.0834356792 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 10 |
Hb_004592_020 |
0.0841374364 |
- |
- |
PREDICTED: non-specific lipid-transfer protein 1-like [Jatropha curcas] |
| 11 |
Hb_018709_010 |
0.08534042 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 12 |
Hb_000589_280 |
0.0875273339 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase 5 [Jatropha curcas] |
| 13 |
Hb_010358_010 |
0.0884402361 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 14 |
Hb_023732_020 |
0.0889837701 |
- |
- |
PREDICTED: fatty acyl-CoA reductase 3-like [Jatropha curcas] |
| 15 |
Hb_002835_020 |
0.0895742297 |
- |
- |
PREDICTED: cytochrome P450 71A1-like [Jatropha curcas] |
| 16 |
Hb_028013_010 |
0.0901574408 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 17 |
Hb_023080_010 |
0.0902847098 |
- |
- |
PREDICTED: probable serine incorporator [Jatropha curcas] |
| 18 |
Hb_000229_080 |
0.0915152715 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 19 |
Hb_004981_020 |
0.0918429938 |
- |
- |
PREDICTED: uncharacterized protein LOC103706119 [Phoenix dactylifera] |
| 20 |
Hb_106528_020 |
0.091849022 |
- |
- |
Cationic peroxidase 2 precursor [Theobroma cacao] |