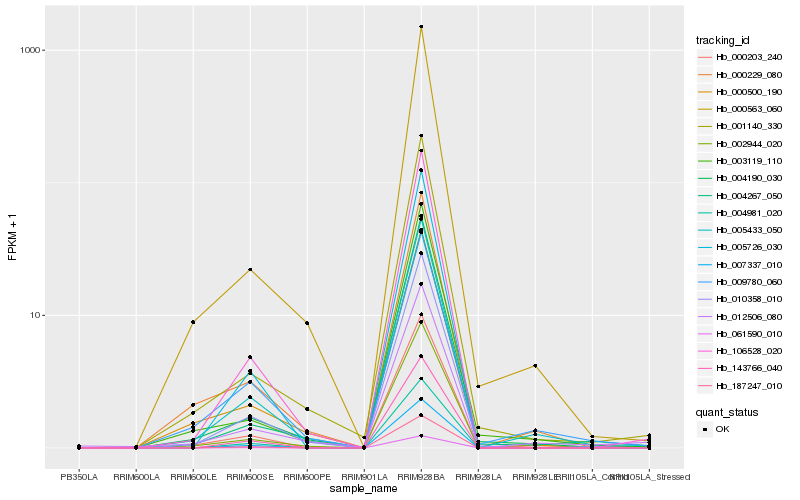
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000203_240 |
0.0 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 2 |
Hb_005433_050 |
0.044060471 |
- |
- |
PREDICTED: uncharacterized protein At1g66480-like [Populus euphratica] |
| 3 |
Hb_002944_020 |
0.0713702217 |
transcription factor |
TF Family: bZIP |
hypothetical protein POPTR_0006s05770g, partial [Populus trichocarpa] |
| 4 |
Hb_000229_080 |
0.0734346741 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 5 |
Hb_106528_020 |
0.0754637071 |
- |
- |
Cationic peroxidase 2 precursor [Theobroma cacao] |
| 6 |
Hb_004190_030 |
0.0788235871 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 7 |
Hb_000563_060 |
0.0794766345 |
- |
- |
PREDICTED: uncharacterized protein LOC100306381 isoform X1 [Glycine max] |
| 8 |
Hb_010358_010 |
0.0803119305 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 9 |
Hb_012506_080 |
0.0803303787 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 10 |
Hb_004267_050 |
0.0805061889 |
- |
- |
hypothetical protein JCGZ_04357 [Jatropha curcas] |
| 11 |
Hb_003119_110 |
0.0815442653 |
- |
- |
PREDICTED: feruloyl CoA ortho-hydroxylase 2-like [Jatropha curcas] |
| 12 |
Hb_009780_060 |
0.0830624862 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranyl geranyl pyrophosphate synthase, putative [Ricinus communis] |
| 13 |
Hb_187247_010 |
0.0832149673 |
- |
- |
PREDICTED: cytochrome P450 86B1-like [Jatropha curcas] |
| 14 |
Hb_061590_010 |
0.0833673273 |
- |
- |
hypothetical protein JCGZ_23475 [Jatropha curcas] |
| 15 |
Hb_005726_030 |
0.0834911846 |
- |
- |
hypothetical protein CISIN_1g020511mg [Citrus sinensis] |
| 16 |
Hb_001140_330 |
0.084371627 |
- |
- |
PREDICTED: uncharacterized protein LOC105142675 [Populus euphratica] |
| 17 |
Hb_000500_190 |
0.0845942077 |
- |
- |
hypothetical protein JCGZ_21655 [Jatropha curcas] |
| 18 |
Hb_007337_010 |
0.0847356039 |
- |
- |
PREDICTED: putative receptor protein kinase ZmPK1 [Jatropha curcas] |
| 19 |
Hb_143766_040 |
0.0850350174 |
- |
- |
PREDICTED: probable caffeoyl-CoA O-methyltransferase At4g26220 [Jatropha curcas] |
| 20 |
Hb_004981_020 |
0.085707868 |
- |
- |
PREDICTED: uncharacterized protein LOC103706119 [Phoenix dactylifera] |