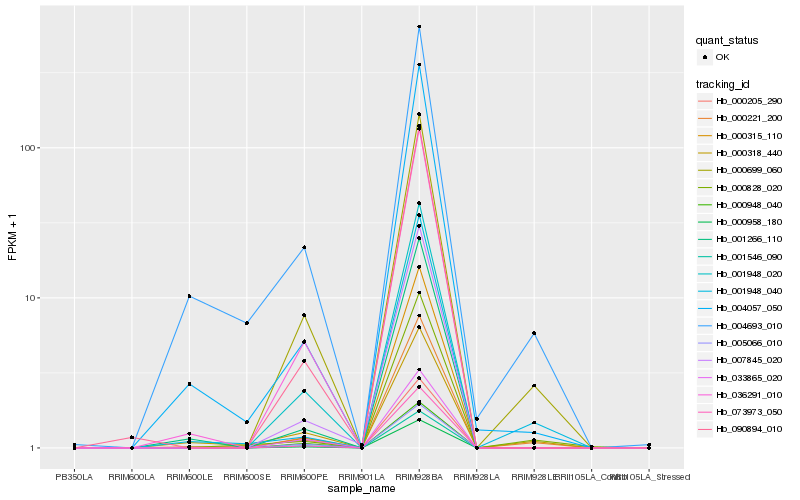
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000699_060 |
0.0 |
- |
- |
hypothetical protein POPTR_0019s03180g [Populus trichocarpa] |
| 2 |
Hb_001546_090 |
0.0679773098 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB114-like [Jatropha curcas] |
| 3 |
Hb_001948_020 |
0.0691580002 |
- |
- |
- |
| 4 |
Hb_000221_200 |
0.0732744956 |
- |
- |
hypothetical protein JCGZ_15819 [Jatropha curcas] |
| 5 |
Hb_036291_010 |
0.0766394429 |
- |
- |
PREDICTED: alpha carbonic anhydrase 7-like [Jatropha curcas] |
| 6 |
Hb_033865_020 |
0.0769350779 |
- |
- |
- |
| 7 |
Hb_005066_010 |
0.0788285595 |
- |
- |
FAD-binding domain-containing family protein [Populus trichocarpa] |
| 8 |
Hb_073973_050 |
0.0835627386 |
transcription factor |
TF Family: GRAS |
PREDICTED: nodulation-signaling pathway 2 protein [Jatropha curcas] |
| 9 |
Hb_000948_040 |
0.085244962 |
- |
- |
PREDICTED: germin-like protein subfamily 1 member 13 [Jatropha curcas] |
| 10 |
Hb_090894_010 |
0.086311354 |
- |
- |
PREDICTED: uncharacterized protein LOC105649214 [Jatropha curcas] |
| 11 |
Hb_007845_020 |
0.0912366831 |
- |
- |
PREDICTED: alpha carbonic anhydrase 7-like [Jatropha curcas] |
| 12 |
Hb_000828_020 |
0.0991860023 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 13 |
Hb_000315_110 |
0.1041878134 |
- |
- |
PREDICTED: gibberellin 3-beta-dioxygenase 1-like [Jatropha curcas] |
| 14 |
Hb_000205_290 |
0.104240925 |
- |
- |
- |
| 15 |
Hb_004693_010 |
0.1093830399 |
- |
- |
hypothetical protein JCGZ_02321 [Jatropha curcas] |
| 16 |
Hb_001266_110 |
0.1097950492 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_004057_050 |
0.1100495626 |
- |
- |
PREDICTED: cytochrome P450 76A1-like [Jatropha curcas] |
| 18 |
Hb_000958_180 |
0.1119396083 |
- |
- |
hypothetical protein CISIN_1g043061mg, partial [Citrus sinensis] |
| 19 |
Hb_000318_440 |
0.1121685081 |
- |
- |
PREDICTED: allene oxide synthase 2-like [Jatropha curcas] |
| 20 |
Hb_001948_040 |
0.1140939744 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranyl geranyl pyrophosphate synthase, putative [Ricinus communis] |