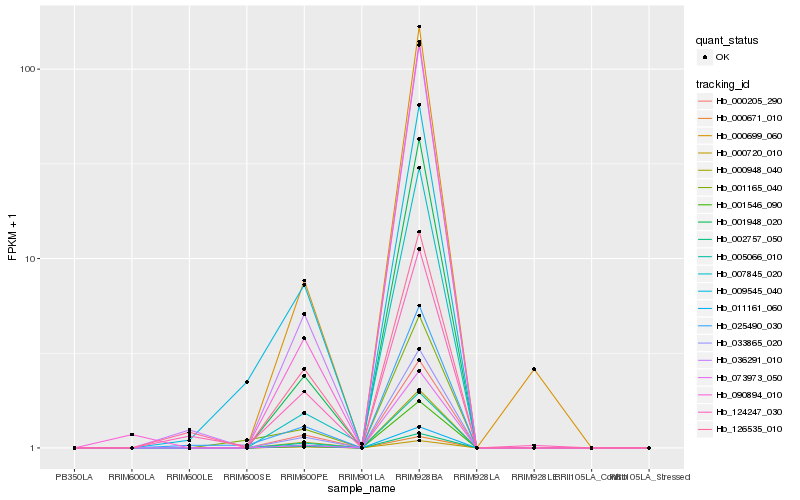
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000948_040 |
0.0 |
- |
- |
PREDICTED: germin-like protein subfamily 1 member 13 [Jatropha curcas] |
| 2 |
Hb_005066_010 |
0.011550301 |
- |
- |
FAD-binding domain-containing family protein [Populus trichocarpa] |
| 3 |
Hb_033865_020 |
0.0154230244 |
- |
- |
- |
| 4 |
Hb_000205_290 |
0.0278767534 |
- |
- |
- |
| 5 |
Hb_002757_050 |
0.0437651112 |
- |
- |
PREDICTED: O-acyltransferase WSD1-like isoform X1 [Populus euphratica] |
| 6 |
Hb_001546_090 |
0.0465611227 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB114-like [Jatropha curcas] |
| 7 |
Hb_000671_010 |
0.0553273238 |
- |
- |
PREDICTED: 7-deoxyloganetin glucosyltransferase-like [Jatropha curcas] |
| 8 |
Hb_011161_060 |
0.0584983778 |
transcription factor |
TF Family: GRAS |
PREDICTED: nodulation-signaling pathway 2 protein [Jatropha curcas] |
| 9 |
Hb_001948_020 |
0.0654843789 |
- |
- |
- |
| 10 |
Hb_036291_010 |
0.0774762125 |
- |
- |
PREDICTED: alpha carbonic anhydrase 7-like [Jatropha curcas] |
| 11 |
Hb_025490_030 |
0.0818052996 |
- |
- |
PREDICTED: uncharacterized protein LOC105629177 [Jatropha curcas] |
| 12 |
Hb_000699_060 |
0.085244962 |
- |
- |
hypothetical protein POPTR_0019s03180g [Populus trichocarpa] |
| 13 |
Hb_000720_010 |
0.0860565735 |
- |
- |
Lignin-forming anionic peroxidase precursor, putative [Ricinus communis] |
| 14 |
Hb_073973_050 |
0.0997512254 |
transcription factor |
TF Family: GRAS |
PREDICTED: nodulation-signaling pathway 2 protein [Jatropha curcas] |
| 15 |
Hb_090894_010 |
0.1013386057 |
- |
- |
PREDICTED: uncharacterized protein LOC105649214 [Jatropha curcas] |
| 16 |
Hb_124247_030 |
0.1023367099 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
PREDICTED: probable 1-deoxy-D-xylulose-5-phosphate synthase 2, chloroplastic [Jatropha curcas] |
| 17 |
Hb_009545_040 |
0.1039930761 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001165_040 |
0.1076643629 |
- |
- |
hypothetical protein POPTR_0009s10120g [Populus trichocarpa] |
| 19 |
Hb_007845_020 |
0.108221339 |
- |
- |
PREDICTED: alpha carbonic anhydrase 7-like [Jatropha curcas] |
| 20 |
Hb_126535_010 |
0.1095230093 |
- |
- |
hypothetical protein JCGZ_09554 [Jatropha curcas] |