| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
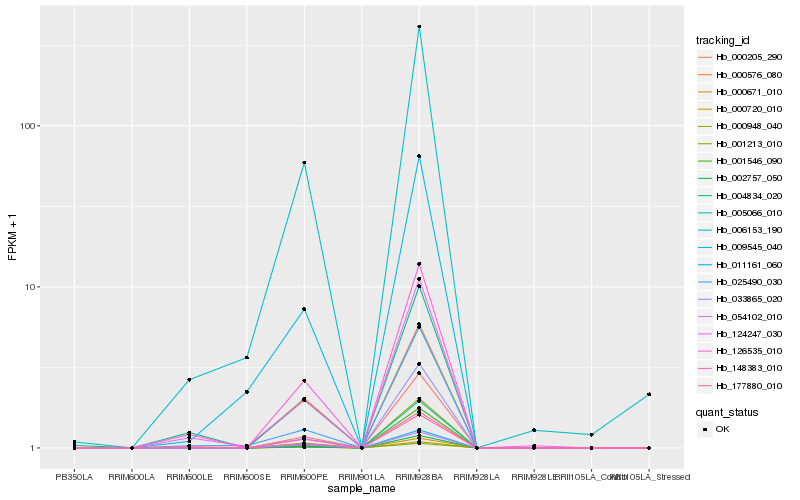
Hb_000671_010 |
0.0 |
- |
- |
PREDICTED: 7-deoxyloganetin glucosyltransferase-like [Jatropha curcas] |
| 2 |
Hb_011161_060 |
0.0031894712 |
transcription factor |
TF Family: GRAS |
PREDICTED: nodulation-signaling pathway 2 protein [Jatropha curcas] |
| 3 |
Hb_002757_050 |
0.0116144754 |
- |
- |
PREDICTED: O-acyltransferase WSD1-like isoform X1 [Populus euphratica] |
| 4 |
Hb_000205_290 |
0.0275330777 |
- |
- |
- |
| 5 |
Hb_000720_010 |
0.0309733693 |
- |
- |
Lignin-forming anionic peroxidase precursor, putative [Ricinus communis] |
| 6 |
Hb_000948_040 |
0.0553273238 |
- |
- |
PREDICTED: germin-like protein subfamily 1 member 13 [Jatropha curcas] |
| 7 |
Hb_001213_010 |
0.0608765987 |
- |
- |
PREDICTED: laccase-7 [Jatropha curcas] |
| 8 |
Hb_054102_010 |
0.0666406127 |
- |
- |
abhydrolase domain containing, putative [Ricinus communis] |
| 9 |
Hb_005066_010 |
0.066784802 |
- |
- |
FAD-binding domain-containing family protein [Populus trichocarpa] |
| 10 |
Hb_033865_020 |
0.0706178056 |
- |
- |
- |
| 11 |
Hb_148383_010 |
0.0793678851 |
- |
- |
PREDICTED: protein YLS9 [Jatropha curcas] |
| 12 |
Hb_006153_190 |
0.0852292131 |
- |
- |
PREDICTED: 14 kDa proline-rich protein DC2.15-like [Gossypium raimondii] |
| 13 |
Hb_126535_010 |
0.0859807588 |
- |
- |
hypothetical protein JCGZ_09554 [Jatropha curcas] |
| 14 |
Hb_000576_080 |
0.0908874133 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_124247_030 |
0.0977595846 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
PREDICTED: probable 1-deoxy-D-xylulose-5-phosphate synthase 2, chloroplastic [Jatropha curcas] |
| 16 |
Hb_009545_040 |
0.0982125875 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001546_090 |
0.1012596299 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB114-like [Jatropha curcas] |
| 18 |
Hb_025490_030 |
0.1030010193 |
- |
- |
PREDICTED: uncharacterized protein LOC105629177 [Jatropha curcas] |
| 19 |
Hb_177880_010 |
0.1171437724 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 20 |
Hb_004834_020 |
0.1175225042 |
- |
- |
major allergen Pru ar 1-like [Jatropha curcas] |