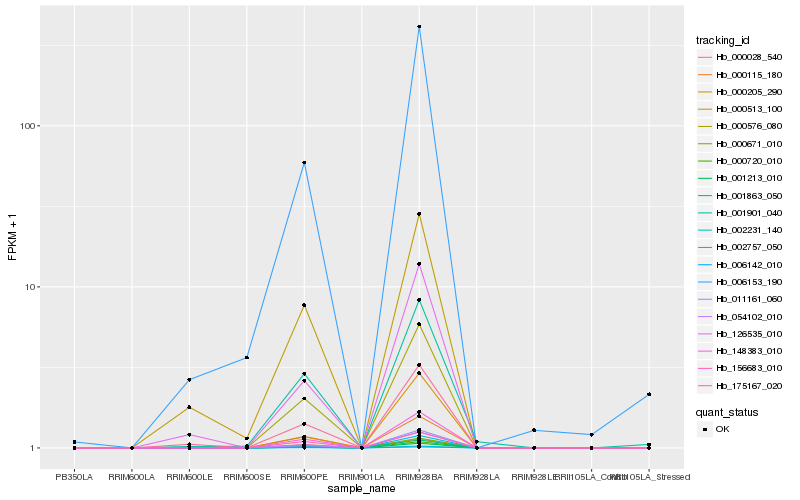
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000576_080 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_148383_010 |
0.0116073244 |
- |
- |
PREDICTED: protein YLS9 [Jatropha curcas] |
| 3 |
Hb_054102_010 |
0.0244060976 |
- |
- |
abhydrolase domain containing, putative [Ricinus communis] |
| 4 |
Hb_001213_010 |
0.0301930964 |
- |
- |
PREDICTED: laccase-7 [Jatropha curcas] |
| 5 |
Hb_000720_010 |
0.0601111931 |
- |
- |
Lignin-forming anionic peroxidase precursor, putative [Ricinus communis] |
| 6 |
Hb_000115_180 |
0.0656735193 |
- |
- |
hypothetical protein RCOM_1308310 [Ricinus communis] |
| 7 |
Hb_001863_050 |
0.0690869201 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 8 [Jatropha curcas] |
| 8 |
Hb_006142_010 |
0.0753556201 |
- |
- |
hypothetical protein CICLE_v10013815mg, partial [Citrus clementina] |
| 9 |
Hb_011161_060 |
0.0877294707 |
transcription factor |
TF Family: GRAS |
PREDICTED: nodulation-signaling pathway 2 protein [Jatropha curcas] |
| 10 |
Hb_000671_010 |
0.0908874133 |
- |
- |
PREDICTED: 7-deoxyloganetin glucosyltransferase-like [Jatropha curcas] |
| 11 |
Hb_002231_140 |
0.0968785751 |
- |
- |
PREDICTED: uncharacterized protein LOC105634453 [Jatropha curcas] |
| 12 |
Hb_000028_540 |
0.0990001587 |
- |
- |
hypothetical protein POPTR_0012s03720g [Populus trichocarpa] |
| 13 |
Hb_006153_190 |
0.1001762183 |
- |
- |
PREDICTED: 14 kDa proline-rich protein DC2.15-like [Gossypium raimondii] |
| 14 |
Hb_156683_010 |
0.1009588012 |
- |
- |
PREDICTED: uncharacterized protein At5g65660-like [Jatropha curcas] |
| 15 |
Hb_002757_050 |
0.1023633501 |
- |
- |
PREDICTED: O-acyltransferase WSD1-like isoform X1 [Populus euphratica] |
| 16 |
Hb_175167_020 |
0.1025817579 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001901_040 |
0.1066269155 |
- |
- |
hypothetical protein CARUB_v10027638mg [Capsella rubella] |
| 18 |
Hb_126535_010 |
0.1141220178 |
- |
- |
hypothetical protein JCGZ_09554 [Jatropha curcas] |
| 19 |
Hb_000205_290 |
0.1180277518 |
- |
- |
- |
| 20 |
Hb_000513_100 |
0.1184011301 |
- |
- |
germin-like protein GLP, partial [Manihot esculenta] |