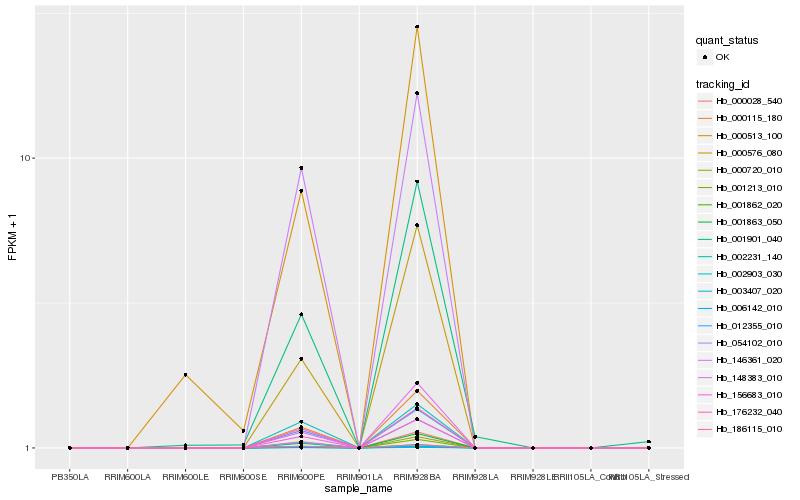
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001863_050 |
0.0 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 8 [Jatropha curcas] |
| 2 |
Hb_000115_180 |
0.0034238798 |
- |
- |
hypothetical protein RCOM_1308310 [Ricinus communis] |
| 3 |
Hb_006142_010 |
0.0062904429 |
- |
- |
hypothetical protein CICLE_v10013815mg, partial [Citrus clementina] |
| 4 |
Hb_002231_140 |
0.027911453 |
- |
- |
PREDICTED: uncharacterized protein LOC105634453 [Jatropha curcas] |
| 5 |
Hb_000028_540 |
0.0300445714 |
- |
- |
hypothetical protein POPTR_0012s03720g [Populus trichocarpa] |
| 6 |
Hb_156683_010 |
0.0320141483 |
- |
- |
PREDICTED: uncharacterized protein At5g65660-like [Jatropha curcas] |
| 7 |
Hb_012355_010 |
0.0496441083 |
- |
- |
PREDICTED: probable protein Pop3 [Jatropha curcas] |
| 8 |
Hb_176232_040 |
0.0629390677 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000576_080 |
0.0690869201 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_003407_020 |
0.0793758345 |
- |
- |
hypothetical protein JCGZ_01308 [Jatropha curcas] |
| 11 |
Hb_148383_010 |
0.0806444691 |
- |
- |
PREDICTED: protein YLS9 [Jatropha curcas] |
| 12 |
Hb_001862_020 |
0.0837509683 |
- |
- |
PREDICTED: uncharacterized protein LOC103652604 [Zea mays] |
| 13 |
Hb_054102_010 |
0.0933690602 |
- |
- |
abhydrolase domain containing, putative [Ricinus communis] |
| 14 |
Hb_146361_020 |
0.0967535241 |
- |
- |
PREDICTED: germin-like protein subfamily 1 member 11 [Jatropha curcas] |
| 15 |
Hb_186115_010 |
0.0983661487 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 16 |
Hb_001213_010 |
0.0991155361 |
- |
- |
PREDICTED: laccase-7 [Jatropha curcas] |
| 17 |
Hb_001901_040 |
0.1072303237 |
- |
- |
hypothetical protein CARUB_v10027638mg [Capsella rubella] |
| 18 |
Hb_002903_030 |
0.10886576 |
- |
- |
polyphenol oxidase [Cydonia oblonga] |
| 19 |
Hb_000513_100 |
0.1244286236 |
- |
- |
germin-like protein GLP, partial [Manihot esculenta] |
| 20 |
Hb_000720_010 |
0.1287490346 |
- |
- |
Lignin-forming anionic peroxidase precursor, putative [Ricinus communis] |