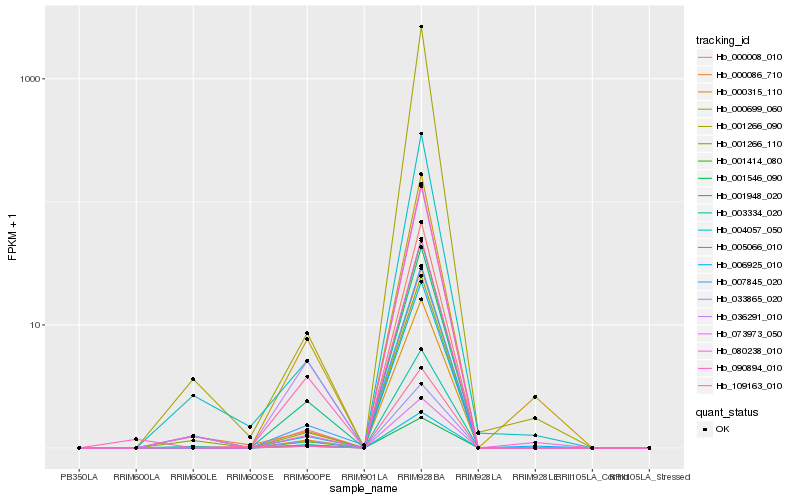
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_090894_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649214 [Jatropha curcas] |
| 2 |
Hb_073973_050 |
0.0249641552 |
transcription factor |
TF Family: GRAS |
PREDICTED: nodulation-signaling pathway 2 protein [Jatropha curcas] |
| 3 |
Hb_007845_020 |
0.039350332 |
- |
- |
PREDICTED: alpha carbonic anhydrase 7-like [Jatropha curcas] |
| 4 |
Hb_000315_110 |
0.0411836909 |
- |
- |
PREDICTED: gibberellin 3-beta-dioxygenase 1-like [Jatropha curcas] |
| 5 |
Hb_001948_020 |
0.0419422211 |
- |
- |
- |
| 6 |
Hb_036291_010 |
0.0476544203 |
- |
- |
PREDICTED: alpha carbonic anhydrase 7-like [Jatropha curcas] |
| 7 |
Hb_001546_090 |
0.0582764449 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB114-like [Jatropha curcas] |
| 8 |
Hb_000008_010 |
0.0590499297 |
- |
- |
PREDICTED: alpha carbonic anhydrase 7-like [Jatropha curcas] |
| 9 |
Hb_001266_110 |
0.0637227209 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_003334_020 |
0.064789309 |
- |
- |
hypothetical protein CISIN_1g041546mg [Citrus sinensis] |
| 11 |
Hb_004057_050 |
0.0721919244 |
- |
- |
PREDICTED: cytochrome P450 76A1-like [Jatropha curcas] |
| 12 |
Hb_001414_080 |
0.0778111416 |
- |
- |
PREDICTED: probable S-adenosylmethionine-dependent methyltransferase At5g38100 [Jatropha curcas] |
| 13 |
Hb_000086_710 |
0.079513636 |
- |
- |
PREDICTED: feruloyl CoA ortho-hydroxylase 2-like [Jatropha curcas] |
| 14 |
Hb_001266_090 |
0.0808752978 |
- |
- |
- |
| 15 |
Hb_109163_010 |
0.085714272 |
- |
- |
polyphenol oxidase [Hevea brasiliensis] |
| 16 |
Hb_080238_010 |
0.0859943727 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 17 |
Hb_000699_060 |
0.086311354 |
- |
- |
hypothetical protein POPTR_0019s03180g [Populus trichocarpa] |
| 18 |
Hb_033865_020 |
0.0869197761 |
- |
- |
- |
| 19 |
Hb_006925_010 |
0.0903533081 |
- |
- |
PREDICTED: uncharacterized protein LOC101777526 [Setaria italica] |
| 20 |
Hb_005066_010 |
0.0905338475 |
- |
- |
FAD-binding domain-containing family protein [Populus trichocarpa] |