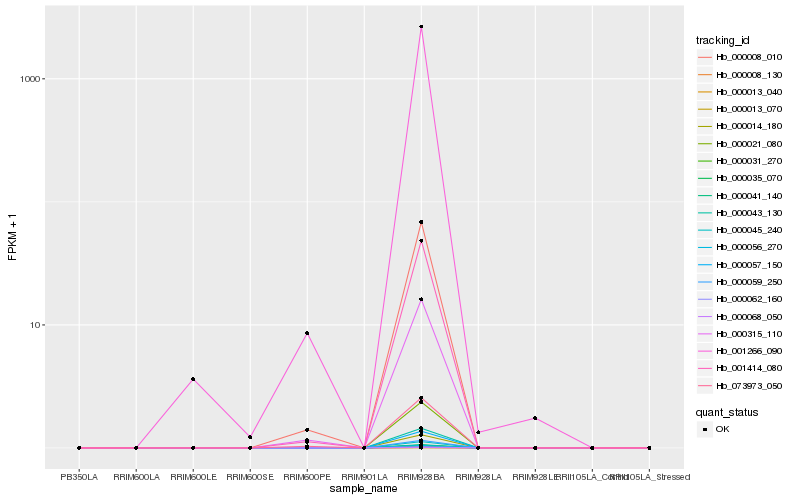
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000008_010 |
0.0 |
- |
- |
PREDICTED: alpha carbonic anhydrase 7-like [Jatropha curcas] |
| 2 |
Hb_000315_110 |
0.0213058412 |
- |
- |
PREDICTED: gibberellin 3-beta-dioxygenase 1-like [Jatropha curcas] |
| 3 |
Hb_001414_080 |
0.0219918627 |
- |
- |
PREDICTED: probable S-adenosylmethionine-dependent methyltransferase At5g38100 [Jatropha curcas] |
| 4 |
Hb_001266_090 |
0.0337770191 |
- |
- |
- |
| 5 |
Hb_073973_050 |
0.0520348166 |
transcription factor |
TF Family: GRAS |
PREDICTED: nodulation-signaling pathway 2 protein [Jatropha curcas] |
| 6 |
Hb_000008_130 |
0.0545506109 |
- |
- |
PREDICTED: uncharacterized protein LOC105630320 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000013_040 |
0.0545506109 |
- |
- |
- |
| 8 |
Hb_000013_070 |
0.0545506109 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 9 |
Hb_000014_180 |
0.0545506109 |
- |
- |
PREDICTED: endo-1,4-beta-xylanase D-like [Fragaria vesca subsp. vesca] |
| 10 |
Hb_000021_080 |
0.0545506109 |
- |
- |
- |
| 11 |
Hb_000031_270 |
0.0545506109 |
transcription factor |
TF Family: B3 |
DNA binding protein, putative [Ricinus communis] |
| 12 |
Hb_000035_070 |
0.0545506109 |
- |
- |
PREDICTED: uncharacterized protein LOC104814513 [Tarenaya hassleriana] |
| 13 |
Hb_000041_140 |
0.0545506109 |
- |
- |
- |
| 14 |
Hb_000043_130 |
0.0545506109 |
- |
- |
- |
| 15 |
Hb_000045_240 |
0.0545506109 |
- |
- |
- |
| 16 |
Hb_000056_270 |
0.0545506109 |
- |
- |
Origin recognition complex subunit, putative [Ricinus communis] |
| 17 |
Hb_000057_150 |
0.0545506109 |
- |
- |
hypothetical protein EUTSA_v10009202mg [Eutrema salsugineum] |
| 18 |
Hb_000059_250 |
0.0545506109 |
- |
- |
PREDICTED: 40S ribosomal protein S8 [Jatropha curcas] |
| 19 |
Hb_000062_160 |
0.0545506109 |
- |
- |
3'-N-debenzoyl-2'-deoxytaxol N-benzoyltransferase, putative [Ricinus communis] |
| 20 |
Hb_000068_050 |
0.0545506109 |
- |
- |
PREDICTED: uncharacterized protein LOC105774125 [Gossypium raimondii] |