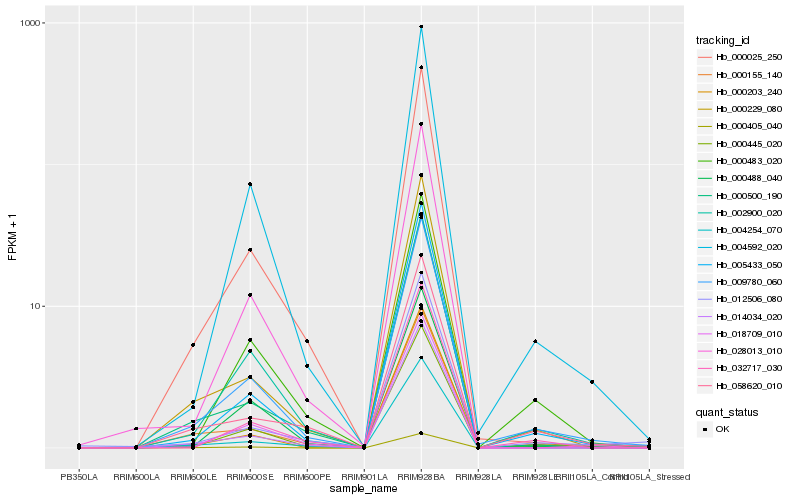
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009780_060 |
0.0 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranyl geranyl pyrophosphate synthase, putative [Ricinus communis] |
| 2 |
Hb_002900_020 |
0.0679952488 |
- |
- |
hypothetical protein POPTR_0008s06270g [Populus trichocarpa] |
| 3 |
Hb_004592_020 |
0.074362415 |
- |
- |
PREDICTED: non-specific lipid-transfer protein 1-like [Jatropha curcas] |
| 4 |
Hb_005433_050 |
0.0768004365 |
- |
- |
PREDICTED: uncharacterized protein At1g66480-like [Populus euphratica] |
| 5 |
Hb_000229_080 |
0.0786721504 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 6 |
Hb_000445_020 |
0.0787673325 |
- |
- |
GABA-specific permease, putative [Ricinus communis] |
| 7 |
Hb_000025_250 |
0.0797681155 |
- |
- |
PREDICTED: flavonoid 3',5'-hydroxylase 2-like [Jatropha curcas] |
| 8 |
Hb_000203_240 |
0.0830624862 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 9 |
Hb_058620_010 |
0.0838806331 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 10 |
Hb_000155_140 |
0.0843286194 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 11 |
Hb_000500_190 |
0.0894384241 |
- |
- |
hypothetical protein JCGZ_21655 [Jatropha curcas] |
| 12 |
Hb_000483_020 |
0.090573685 |
- |
- |
PREDICTED: 7-deoxyloganetic acid glucosyltransferase-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_014034_020 |
0.0912651347 |
- |
- |
hypothetical protein POPTR_0075s00200g, partial [Populus trichocarpa] |
| 14 |
Hb_032717_030 |
0.0921561055 |
- |
- |
PREDICTED: CASP-like protein 1B2 [Jatropha curcas] |
| 15 |
Hb_000488_040 |
0.0928868403 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 16 |
Hb_004254_070 |
0.0943355322 |
transcription factor |
TF Family: NAC |
hypothetical protein POPTR_0015s03700g [Populus trichocarpa] |
| 17 |
Hb_018709_010 |
0.0951104602 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 18 |
Hb_012506_080 |
0.0955055766 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 19 |
Hb_028013_010 |
0.096865689 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 20 |
Hb_000405_040 |
0.0971258349 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |